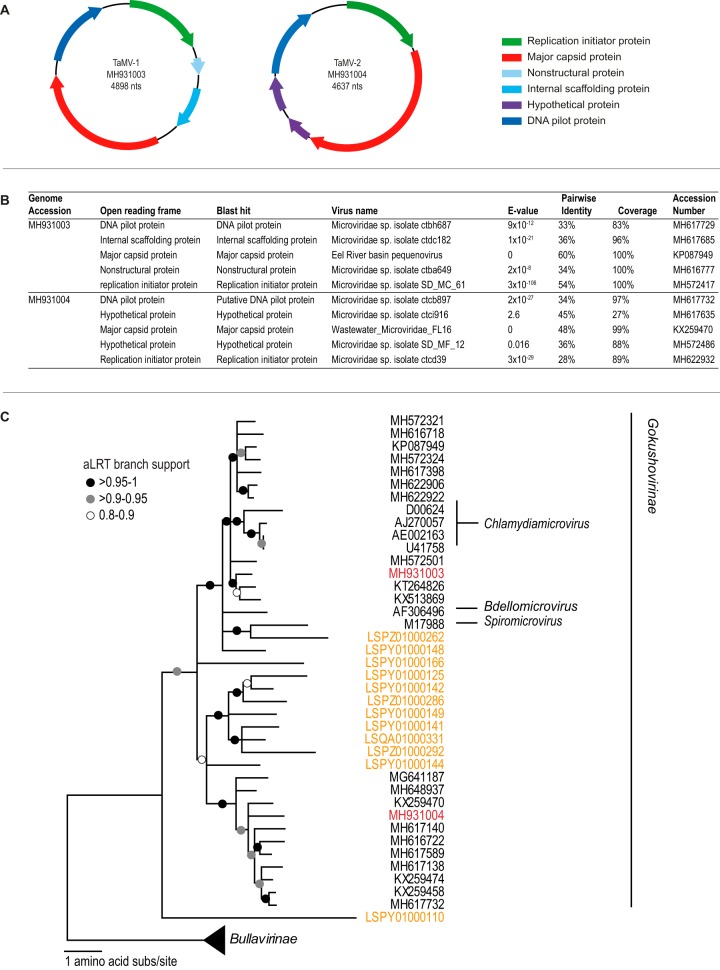
FIG 1.
(A) Genome organization of termite-associated microvirus-1 (replication initiator protein, 882 nucleotides [nt]; nonstructural protein, 276 nt; internal scaffolding protein, 468 nt; major capsid protein, 1,704 nt; and DNA pilot protein, 837 nt) and termite-associated microvirus-2 (replication initiator protein, 1,017 nt; major capsid protein, 1,608 nt; hypothetical proteins, 339 and 417 nt; and DNA pilot protein, 768 nt). (B) Summary of the best BLASTp results for each ORF of TaMV-1 and TaMV-2. (C) Maximum likelihood phylogenetic tree of the MCP amino acid sequences and the pairwise identities of the MCP of most closely related Gokushovirinae members, those from termite reported by Tikhe and Husseneder (5), and those from this study. Numbers in red are MCP sequences from this study, and numbers in orange are MCP sequences identified in termites by Tikhe and Husseneder (5). The maximum likelihood phylogenetic trees were inferred with PhyML (17) with the RtRev+F+G substitution model and with approximate likelihood ration test (aLRT) branch support.