FIGURE 6.
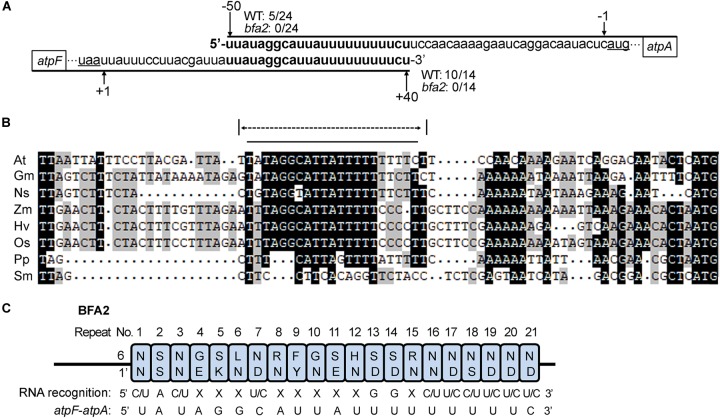
Position of the BFA2-dependent RNA termini. (A) Overlapping transcript termini in the atpF/A intergenic region. Nucleotide positions relative to the start codon of atpA (-50 and -1) and stop codon of atpF (+40 and +1) are shown above and below the sequences, respectively. The numbers of 5/24, 0/24, 10/14, and 0/14 represent the ratio of the clones with a 5′ or 3′ end positions from the WT or bfa2 samples in the cRT-PCR analyses (Supplementary Figure S5 and Supplementary Table S1). (B) Alignment of the atpF/A intergenic regions (from the stop codon of atpF to the start codon of atpA). The overlapping transcript termini in the atpF/A intergenic regions and the sequence predicted for BFA2 binding are indicated by the dotted and black lines, respectively. Nucleotides U are indicated by T. At, Arabidopsis thaliana; Gm, Glycine max; Os, Oryza sativa; Zm, Zea mays; Ns, Nicotiana sylvestris; Hv, Hordeum vulgare; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii. (C) Prediction of the nucleotide-binding site for BFA2. The residues that determine nucleotide-binding specificity (residues 6 and 1’ in Supplementary Figure S3) in each PPR motif were extracted according to Barkan et al. (2012). The overlapping sequence in the atpF/A intergenic RNA (from the second nucleotide) is shown.