Abstract
Anthropologists have a lengthy history using cranial nonmetric traits to assess biological distances between populations. These concepts were adopted by forensic anthropologists to estimate ancestry at the individual level using population-based human variation. However, this method of estimation must adhere to the Daubert guidelines to be applied in the modern court system. To that end, a set of cranial macromorphoscopic traits have been proffered to generate a methodological approach to the estimation of ancestry that can be empirically tested and verified. Additionally, various statistical frameworks, parametric and nonparametric, have been employed to calculate error rates and provide probabilistic statements of the strength of these estimations. Recent studies have further advanced the reliability and validity of macromorphoscopic trait analysis through reference datasets, validation studies, and demonstrable exploration of the complex population histories impacting human variation between and within populations worldwide.
Keywords: Forensic pathology, Forensic anthropology, Ancestry estimation, Macromorphoscopic traits, Classification statistics, Daubert
Introduction
Ancestry estimation from human skeletal remains is one of the primary objectives of the forensic anthropologist when establishing a biological profile for unidentified human skeletal remains. Such estimations can be accomplished through various methods, including cranial metrics (1, 2), dental metrics (3, 4), dental morphology (5, 6), and macromorphoscopic traits (7). Cranial metric analysis is generally the most utilized method for ancestry estimation; however, this method is of limited use when skeletal remains are fragmented or poorly preserved (8). Macromorphoscopic traits can be coupled with cranial metrics to strengthen ancestry estimations or they may provide an alternative method to classify ancestry when cranial metric data cannot be collected.
Macromorphoscopic traits are slight variations in cranial form (7), which capture some aspect of cranial morphology through an assessment of 1) a bone's shape (e.g., nasal bone structure), 2) a bony feature morphology (e.g., inferior nasal aperture morphology), 3) a suture's shape (e.g., zygomaticomaxillary suture), 4) the presence or absence of a particular feature (e.g., postbregmatic depression), and 5) a bony prominence or protrusion (e.g., anterior nasal spine) (7). Cranial morphology as a tool to discriminate between groups of humans is not a new approach, but is rooted in the works of 18th and 19th century natural historians and biologists. For example, Linnaeus created a taxonomy of four human subspecies (Homo sapiens amercianus, africanus, asiaticus, and europaeus) based on morphological variation between crania (9). While others built upon the work of Linnaeus, modern forensic anthropologists rely on procedures developed at least initially by E. A. Hooton (1887-1954) at Harvard University. Hooton focused the majority of his studies on human variation using cranial metrics and cranial nonmetric traits to distinguish between so-called “races” of humans.
Hooton's acceptance of the biological validity of discrete packages of humans led him to believe adaptive traits—those that respond to environmental influences—should be avoided so analyses could focus on variation specific to each biological race (10). Hooton's methodology to racial classification from cranial morphology resulted in a long-standing acceptance of the typological approach to ancestry estimation in forensic anthropology (8). Until very recently, most forensic anthropologists viewed cranial nonmetric traits (i.e., macromorphoscopic traits) as expressions of a character state specific to one ancestral group, a method we now refer to as the “trait list approach” (8, 11).
The trait list approach is the result of the Harvard List, a scoring system for traditional cranial morphological traits initially developed by Hooton (12, 13) to determine biological relatedness between populations via archaeologically derived skeletal samples. Today, the trait list approach centers on line drawings and trait tables outlining so-called expected trait values within groups based on typological models of human variation. As conflicting theories on the existence of race developed within science in general and more specifically in anthropology, many anthropologists switched focus from studies of race to population-based studies intending to understand the complexities of human variation without the use of broadly based geographic races.
Throughout the 20th century, forensic anthropologists continued to use the Harvard List scoring system. Unfortunately, the continued reliance on this subjective method results in an approach to ancestry estimation that relies heavily on the experience of the observer and does not account for the total range in human variation due to the typological nature of trait lists. Moreover, the visual assessment of these traits and subsequent comparison to trait lists is usually followed by post hoc trait selection to support a conclusion, an unscientific and unverifiable approach in any science. The trait list approach was unscientific and unverifiable because there was no way to empirically test the method nor could forensic anthropologists calculate an error rate for a final estimation of ancestry (14). Although the trait list approach to the estimation of ancestry has largely fallen out of favor, the method continues to be used in some laboratories today.
Forensic anthropologists still using the trait list approach frequently cite Rhine's study on nonmetric traits (15) to justify using the method in forensic case reports. Rhine documented 45 cranial traits for American black and white, Hispanic, and Amerindian groups (15). However, as he notes, the results are somewhat uncertain due to limited sample sizes and the high occurrence of unexpected trait values within groups (16). In addition to those shortcomings, the trait list approach does not currently meet the Daubert criteria. To satisfy the Daubert requirements, Hefner developed a standardized and more objective framework for scoring macromorphoscopic traits (7). This new methodology is a more rigorous approach to ancestry estimation and provided a starting point for future work.
Discussion
Macromorphoscopic Trait Analysis
The Daubert guidelines underscored the need to produce a more scientific framework for macromorphoscopic trait analysis. Ousley and Hefner coined the term macromorphoscopic traits by noting the differences between the traditional cranial nonmetric traits used in biodistance analysis for population comparisons and the traits used by forensic anthropologists to estimate ancestry for an individual. They also hinted at the potential for trait analysis on a more robust level using statistical analyses (14). Building on that research, Hefner examined eleven macromorphoscopic traits for African, European, Native American, and Asian populations and indirectly recommended several statistical procedures appropriate for classifying unknown crania into an ancestral group (7). Table 1 lists a number of macromorphoscopic traits and their standard abbreviations. Hefner provides illustrations (line drawings) and definitions for these traits (7). His research focused on determining trait frequencies within and between groups, intertrait correlations, and suitable statistical procedures for classifications. Hefner reported classification rates between 84% and 93% depending on the statistical method used (i.e., logistic regression, naïve Bayesian, k-nearest neighbor) and the combination of traits incorporated in an analysis (7).
Table 1.
Macromorphoscopic Traits and Their Standard Abbreviation
| Morphoscopic Trait | Abbreviation |
|---|---|
| Anterior nasal spine | ANS |
| Inferior nasal aperture | INA |
| Interorbital breadth | IOB |
| Malar tubercle | MT |
| Nasal aperture width | NAW |
| Nasal bone contour | NBC |
| Nasal overgrowth | NO |
| Palate shape | PS |
| Postbregmatic depression | PBD |
| Supranasal suture | SPS |
| Transverse palatine suture | TPS |
| Zygomaticomaxillary suture | ZS |
Applied statistical methods are lacking from the earlier work of Hefner and colleagues. Recently, Hefner (17) and Hefner and Ousley (18) reported that using fewer traits may provide more accurate classifications. By narrowing the list of traits used in an analysis, the post-hoc nature of trait selection is mitigated. The traits suggested by these researchers were selected based on previous research identifying their utility for classifying unknown ancestry between American black, American white, and Hispanic groups and through stepwise variable selection procedures in multivariate statistical analyses (17). We will focus herein on seven of the more commonly employed traits. A description of each is provided below. Figures 1–3 provide the general location of each trait on a diagrammatic cranium. Figures corresponding to the descriptions below are available from Hefner (7, 17).
Figure 1.
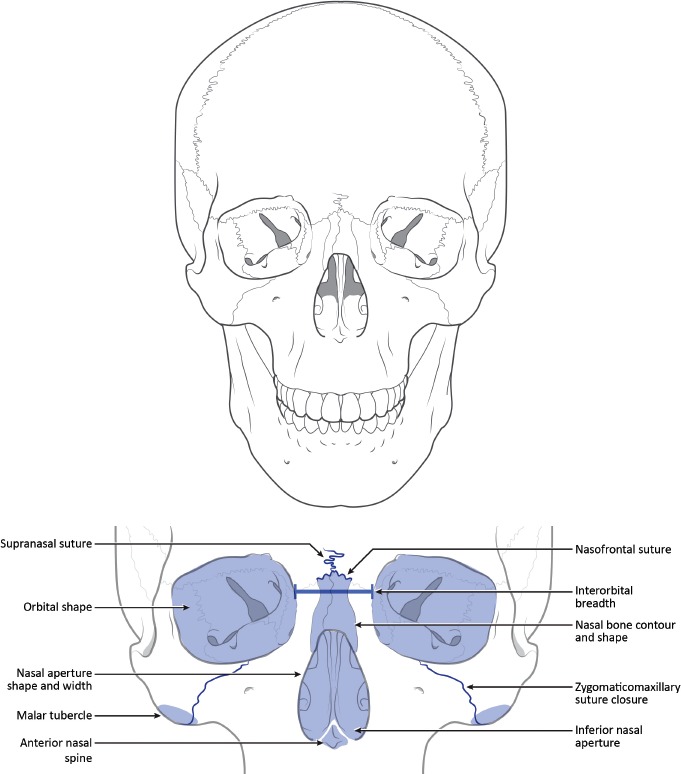
Anterior views of a cranium to demonstrate approximate location of macromorphoscopic traits. Drawn under contract with professional medical illustrator Diana Kryski.
Figure 3.
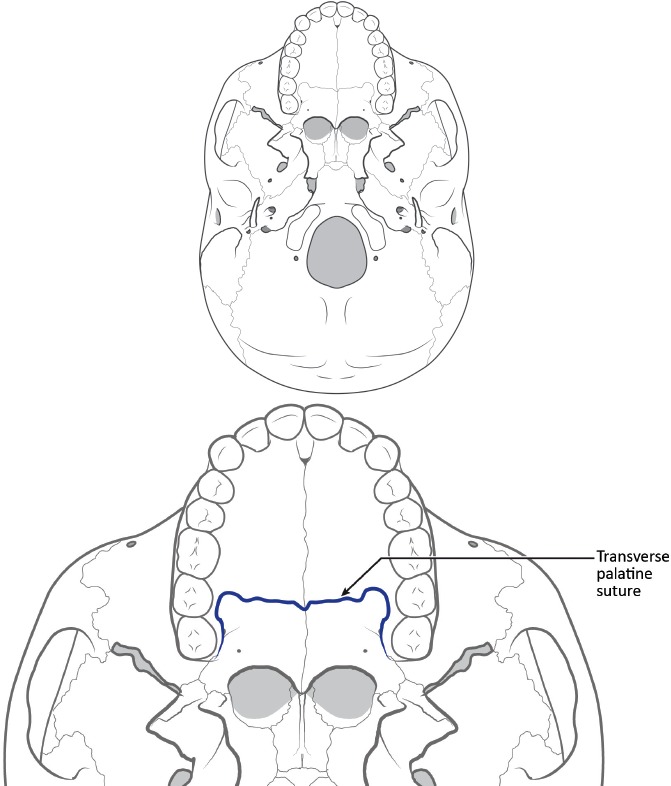
Inferior views of a cranium to demonstrate approximate location of macromorphoscopic traits. Drawn under contract with professional medical illustrator Diana Kryski.
Figure 2.
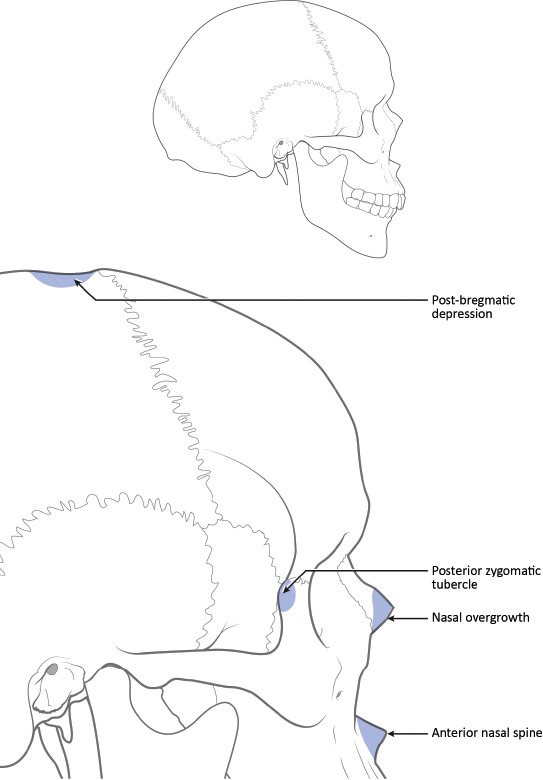
Lateral views of a cranium to demonstrate approximate location of macromorphoscopic traits. Drawn under contract with professional medical illustrator Diana Kryski.
Macromorphoscopic Traits
The anterior nasal spine (ANS) is a protrusion of bone extending forward at the base of the nasal aperture. In some cases the anterior nasal spine is quite small; in others the anterior nasal spine dominates the vertical maxilla. Because of the fragility of the anterior nasal spine and the position of this trait on the face, taphonomic factors and pathological responses to disease can alter the projection of the spine. Those crania exhibiting trauma, pathology (including alveolar resorption), or postmortem damage to the overall inferior nasal margin should be excluded from the analysis. To score the anterior nasal spine, view the cranium laterally. Anterior nasal spine is scored progressively as slight, intermediate, and marked: 1) Slight- minimal to no projection of the anterior nasal spine beyond the inferior nasal aperture; 2) Intermediate- a moderate projection of the anterior nasal spine beyond the inferior nasal aperture; 3) Marked- a pronounced projection of the anterior nasal spine beyond the inferior nasal aperture.
Inferior nasal morphology (INA) is the most inferior portion of the nasal aperture, which, when combined with the lateral alae, constitutes the transition from the nasal floor to the vertical portion of the maxillae, superior to the anterior dentition. Inferior nasal aperture is an assessment of the shape of the inferior border of the nasal aperture just lateral to the anterior nasal spine. The morphology of INA ranges from an inferior slope with no delineation of the inferior border (guttering), to a sharp and vertical ridge of bone (nasal sill). The five character states of INA should be scored following these guidelines: 1) Guttering- a pronounced sloping of the nasal floor beginning within the nasal cavity and terminating on the vertical surface of the maxilla, producing a smooth transition (this morphology is can be differentiated from INA 2) by noting the more posterior origin and greater slope of INA 1; 2) Partial Guttering- a moderate sloping of the nasal aperture beginning more anteriorly than in INA 1, and with more angulation at the exit of the nasal opening; 3) Straight- a straight transition from nasal floor to the vertical maxilla with no intervening projection of bone, usually angled sharply, although more blunted forms have been observed; 4) Partial Sill- any superior projection of the anterior nasal floor, creating a weak (but present) vertical ridge of bone traversing the inferior nasal border (partial sill); 5) Nasal Sill- a pronounced ridge (sill) obstructing the nasal floor-to-maxilla transition.
Interorbital breadth (IOB) is the distance between the dacrya. As a macromorphoscopic trait, interorbital breadth is assessed relative to the total breadth of the facial skeleton: 1) Narrow, 2) Intermediate, and 3) Wide.
Nasal aperture width (NAW) is the breadth of the nasal aperture relative to the facial skeleton. The width of the nasal aperture is: 1) Narrow, 2) Medium, and 3) Broad.
Nasal bone contour (NBC) is the contour of the mid-facial skeleton in the region where the nasal bones and the frontal process of the maxilla meet. Assess the contour approximately 1 cm below nasion. The following character states are used to assess NBC: 0) low and rounded; 1) an oval contour, with elongated, high, and rounded lateral wall; 2) steep lateral walls with a broad (roughly 7 mm or more) and flat superior surface plateau; 3) steep-sided lateral walls and a narrow superior surface plateau; 4) triangular cross section and no superior surface plateau.
Nasal overgrowth (NO) is the inferior projection of the lateral border of the nasal bones beyond the maxillae in the area of nasale inferious. Assessment of nasal overgrowth does not include anterior bulging of the nasal bones. Observations should be made of the left side if it is undamaged. If the left side is damaged, the right side may be substituted. If both nasal bones are missing or fractured (ante-, peri-, or postmortem), the trait is not scored. Running a finger along the borders of the maxilla and nasal bones near nasale inferious to determine whether a projection is present may be useful. Nasal overgrowth is scored dichotomously as 0) absent or 1) present.
Postbregmatic depression (PBD) is a slight to broad depression along the sagittal suture posterior to bregma that is not the result of pathology or trauma. Observed in the lateral profile, the trait is scored as 0) absent or 1) present.
Analytical Options
There are several tools available for the collection of macromorphoscopic data. The computer program Osteoware (Smithsonian Institution, 2015) includes the eleven macromorphoscopic traits originally assessed by Hefner (7) as well as five additional traits (nasal aperture shape, nasal bone shape, naso-frontal suture, orbital shape, and posterior zygomatic tubercle). This software provides descriptions for each trait expression together with images and a back-end data management system using a relational database. Following data collection, trait scores can then be applied to classification models providing the necessary statistical framework for analysis. Another program, based largely on Osteoware, is Macromorphoscopic Traits 1.61 (MMS), a free-standing beta-version designed for data collection. This program is one part of a data collection effort for the Macromorphoscopic Databank (described in more detail below).
There are a number of appropriate classification statistics for analyzing macromorphoscopic data. Hefner suggests k-nearest neighbor, canonical analysis of principal coordinates (CAP), artificial neural networks, and other non- and semi-parametric methods to classify unidentified individuals (8). Hefner and Ousley outline multiple methods of analysis and provide a novel statistic, the Optimized Summed Scored Attributes, or OSSA, for a dichotomous classification between American blacks and American whites (18). This method relies on the bimodal distribution of optimized macromorphoscopic trait scores to provide an estimation of ancestry. More detailed discussions on these statistical methods are available in Hefner (8) and Hefner and Ousley (18). Several case studies presented below will guide the reader through a typical analytical process, demonstrating trait scoring, a relatively simple analytical process, as well as reporting strategies for casework.
More Recent Advances
Macromorphoscopic traits analysis has recently witnessed expansion and improvement. Two primary lines of research include testing the validity and observer error of Hefner's original study (19, 20,21) and expanding that method to other population groups (22-25). Several important studies are highlighted below.
Klales and Kenyhercz (21) conducted a validation study of Hefner (7). This study adhered to the Daubert guidelines, which require methods to be empirically tested and validated. The authors examined 208 American white and American black individuals from the Hamann-Todd Osteological Collection to determine if classification is successful between the two groups, whether sex impacts classification rates, and assess observer error between experienced and inexperienced observers. Overall, this study supported the findings of Hefner (7) with similar trait expressions between the groups, as well as higher classification rates when pooling male and female samples in the reference collection. The classification accuracies ranged from 73.3% to 86.6% depending on the combination traits used and the statistical procedure applied, with rates falling to 46.7% and 64.3% when sex is included (19). However, the authors found that interobserver error was greater between experienced and inexperienced observers than previously reported by Hefner (7) and L'Abbe et al. (22). Klales and Kenyhercz (21) recommend forensic anthropologists familiarize themselves with macromorphoscopic traits, their various character state expressions, and the trait definitions prior to performing an analysis during casework (19).
The second area of advancement in macromorphoscopic trait research is expansion of these methods to other groups beyond the traditional American populations. Several studies broaden the application of this method to accommodate current populations across the globe. First, the rising Hispanic population in the United States required updated ancestry methods. Recent studies focused on determining which macromorphoscopic traits are most reliable for classifying Hispanic Americans, as well as establishing trait frequencies for reference samples and calculating classification rates for these groups (23, 24). Spradley and Hefner argue that the traditional three-group model (African, European, and Asian or Hispanic or Native American) of ancestry estimation should no longer be used (26). The authors demonstrate it is possible to correctly classify an individual beyond the three-group model when applying Artificial Neural Network statistics (26).
Additionally, researchers in other parts of the world are adapting these methods to approach ancestry estimation in their region. For example, L'Abbe et al. used 13 macromorphoscopic traits to determine if ancestry is correlated with South African social races (white, black, and “colored”) (22). Nine traits from Hefner (7) (NBC, NAW, ANS, INA, NO, MT, IOB, ZS, and TPS) were used in their analysis. Additionally, zygomatic and alveolar projection (27), mandibular torus, and palatine torus (28) were incorporated to correspond to South African forensic case reports using those traits. The authors found significant correlations between all traits, with the exception of the transverse palatine suture shape. Additionally, this study reported seven traits (NO, INA, NAW, ANS, NBC, IOB, and MT) being highly to moderately repeatable in scoring with very little interobserver error, while the remaining six are highly variable and may not be reliable for forensic casework. The six traits with high observer errors are not present in Hefner's suggested seven-trait suite (17).
Rather than focusing on classification of social race using macromorphoscopic traits, Ratliff used trait variation between Pacific Island groups (island Southeast Asia, Melanesia, and Polynesia) and mainland Asia (China and Japan) to document trait variation arising from geographic boundaries and inhibited gene flow (25). She found that the Pacific Island groups were more related morphologically and differentially expressed macromorphoscopic traits compared to mainland Asia. All traits examined were statistically different between these groups with the exception of postbregmatic depression and nasal overgrowth (24). Regional studies such as these are important for identifying the degree of human variation between populations.
Researchers are also combining cranial macromorphoscopic trait data with other ancestry estimation methods, such as genetic markers (i.e., mitochondrial DNA, Y-chromosomes, and single nucleotide polymorphisms) (29) and cranial metric data (30). By combining multiple methods of ancestry estimation, forensic anthropologists have multiple lines of evidence to support their classifications and increase the empirical nature used to establish a biological profile of an unidentified individual.
One final notable advance in macromorphoscopic trait analysis is the introduction of the Macromorphoscopic Databank, or MaMD, a National Institute of Justice funded project (NIJ 2015-DN-BX-K012) designed for the collection and analysis of macromorphoscopic data from around the world. The MaMD will serve two primary purposes: 1) as a repository for macromorphoscopic trait data; and 2) as reference data for an analytical program designed for ancestry estimations using these traits. Currently, MaMD contains data for over 2400 individuals from around the world. Trait scores for all of the aforementioned traits are included along with an additional trait (palate shape) recently added to the Macromorphoscopic Module (MMS). The data collection program and the reference data will be available to researchers following completion of the NIJ project.
Case Studies
The following case studies outline the analysis of macromorphoscopic traits for the estimation of ancestry. These examples are drawn from casework at several laboratories. Although full biological profiles were conducted, only the ancestry estimation is discussed below. Case numbers and other identifying information have been redacted. All analyses were conducted using reference data from MaMD.
Image 1 presents the cranial remains of a previously unidentified middle-aged male. Table 2 provides the individual macromorphoscopic trait scores. We utilized five potential groups from the MaMD (American black, American Indian, American white, Asian, and Hispanic) and twelve macromorphoscopic traits (ANS, INA, IOB, MT, NAS, NAW, NBC, NBS, NO, PBD, TPS, and PS) in a CAP classification analysis with stepwise variable selection. Table 3 provides classification statistics for this individual. Overall, the CAP analysis correctly classified 63% of the reference sample using seven traits (ANS, INA, IOB, NAW, NBC, NO, and PBD). Case study 1 is classified as American white (d2 = 5.4; posterior probability = 0.477). However, the classification statistics for Hispanic are also very close (d2 = 5.5; posterior probability = 0.451). These results are seemingly ambiguous between American white and Hispanic; for a forensic anthropology report, the ancestry estimation would best be stated as: “Based on seven macromorphoscopic traits and a 6-group CAP analysis, this individual is most likely American white, although Hispanic ancestry cannot be ruled out.” Of course, if craniometric data are also available, then the final assessment of ancestry may rely on both methods to further refine the overall statement on ancestry. The individual in case study 1 was later identified as a white male.
Table 2.
Macromorphoscopic Trait Scores for the Two Case Studies
| Case Study 1 |
Case Study 2 |
||
|---|---|---|---|
| Trait | Score | Trait | Score |
| ANS | 3 | ANS | 1 |
| INA | 4 | INA | 1 |
| IOB | 1 | IOB | 2 |
| MT | 1 | MT | 1 |
| NAS | 1 | NAS | 3 |
| NAW | 1 | NAW | 2 |
| NBC | 4 | NBC | 1 |
| NBS | 3 | NBS | – |
| NO | 1 | NO | 0 |
| PBD | 0 | PBD | 1 |
| TPS | 3 | TPS | 2 |
| PS | 3 | PS | 3 |
Table 3.
Classification Data for Case Study 1
| Group | Classification | d2 | Posterior Probability |
|---|---|---|---|
| American white | AMW | 5.4 | 0.628 |
| Hispanic | 5.5 | 0.615 | |
| Amerindian | 9.7 | 0.056 | |
| Asian | 12.3 | 0.015 | |
| American black | 18.5 | 0.001 |
Image 1.
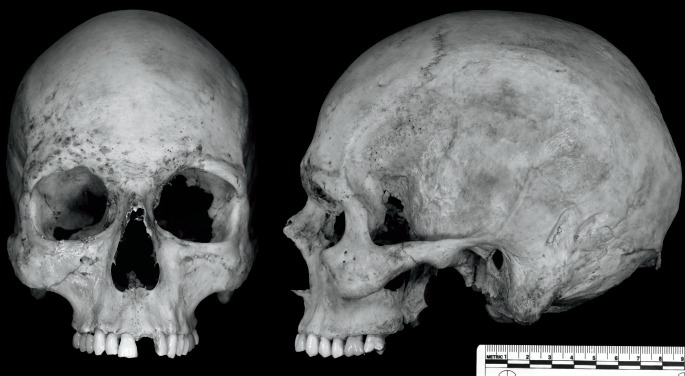
Two views (anterior and lateral) of the cranium from case study 1.
The cranial remains of case study 2 are provided in Image 2. These remains represent a previously unidentified young adult female. See Table 2 for the individual macromorphoscopic trait scores. For this analysis, we utilized the five potential groups and eleven macromorphoscopic traits in a CAP classification analysis with stepwise variable selection. Table 4 provides classification statistics for this individual. Overall, the CAP analysis correctly classified 65% of the reference sample using seven traits (ANS, INA, IOB, NAW, NBC, NO, and PBD). Case study 2 is classified as American black (d2 = 5.6; posterior probability = 0.783). The next closest group is Asian (d2 = 9.2; posterior probability = 0.127). For a forensic anthropology report, this ancestry estimation would best be stated as: “Based on seven macromorphoscopic traits and a 5-group CAP analysis, this individual is most likely American black.” The inclusion of craniometric data to support this analysis would strengthen the overall statement on ancestry. The individual in case study two was identified as a black female.
Image 2.
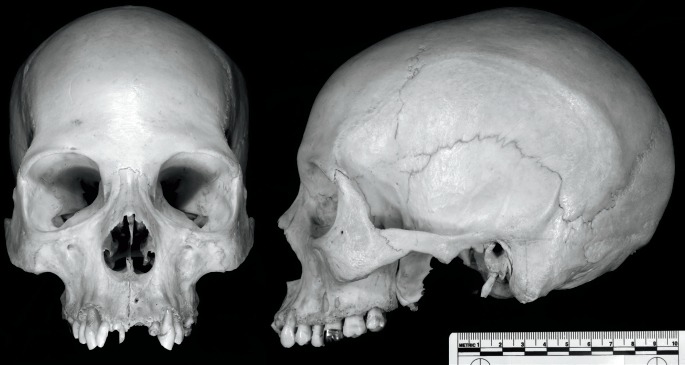
Two views (anterior and lateral) of the cranium from case study 2.
Table 4.
Classification Data for Case Study 2
| Group | Classification | d2 | Posterior Probability |
|---|---|---|---|
| American black | AMB | 5.6 | 0.783 |
| Asian | 9.2 | 0.127 | |
| AmerIndian | 10.9 | 0.055 | |
| American white | 12.8 | 0.021 | |
| Hispanic | 13.6 | 0.014 |
Conclusion
Ancestry estimation is a critical and complex component in the establishment of biological profiles for unidentified human skeletal remains. While cranial morphology has been used to determine ancestry since the early 20th century, those attempts were based on observer experience and typological trait lists (14). Forensic anthropologists now have available more robust and empirically based approaches to macromorphoscopic trait analysis, all designed and developed in an effort to more strictly observe the Daubert guidelines (7). This statistical framework allows the calculation of error rates (How often am I going to be wrong when I estimate ancestry?), in addition to probabilistic statements of an individual belonging to a particular group (How likely am I to be right for this one estimate?). These advances in cranial macromorphoscopic trait analyses should be combined with or, when those data are not possible to collect because of fragmentation or poor preservation, used alternatively to cranial metric analyses.
Forensic anthropologists are continuously improving reference samples and the understanding of population-based human variation to increase the accuracy of ancestry estimation. Recent studies focus on adapting macromorphoscopic trait analyses to accommodate regional judicial system requirements of identifying unknown human remains, such as trait frequencies of prominent ancestral groups or social races unique to a region (22). Historic approaches, such as the three-group (white, black, other) typological model, are being abandoned due complex population histories. The changing demographic structure of the United States is resulting in much greater levels of admixture within the general population. Currently, there is no way to determine admixture from the cranium using these traits, and, until the genetic component of macromorphoscopic traits is better understood and appropriate reference samples are available, this knowledge gap will remain.
However, as the case studies above demonstrate, the switch to multigroup classifications beyond three-group models is not only possible, but works very well. Methodological improvements to—and further refinement, verification, and validation of—the current approaches to the estimation of ancestry will only improve our ability to correctly assess ancestry from human skeletal remains. More accurate and empirically sound biological profiles of unknown human remains increase both the likelihood of identification by law enforcement and the admissibility of anthropological evidence in courts of law.
Acknowledgements
The authors would like to thank the Macromorphoscopic Databank for access to reference data used for the case studies, which includes data from the following institutions and/or individuals: Dawnie Steadman and Lee Meadows Jantz (Department of Anthropology, University of Tennessee, Knoxville, TN), David Hunt (National Museum of Natural History, Smithsonian Institution, Washington, DC), Melody Ratliff (Pacific Islands data), and Bruce Anderson (Pima County Medical Examiner's Office, Tucson, AZ) for providing access to skeletal collections. We are also grateful to the faculty, staff, and graduate students of the Michigan State University, Forensic Anthropology Laboratory for providing the material used for the case studies.
Footnotes
Disclosures
This project was supported by Award No. 2015-DN-BX-K012 from the National Institute of Justice, Office of Justice Programs, U.S. Department of Justice. The authors have indicated that they do not have any other financial relationships to disclose that are relevant to this manuscript
ETHICAL APPROVAL
As per Journal Policies, ethical approval was not required for this manuscript
STATEMENT OF HUMAN AND ANIMAL RIGHTS
This article does not contain any studies conducted with animals or on living human subjects
STATEMENT OF INFORMED CONSENT
No identifiable personal data were presented in this manuscsript
DISCLOSURES & DECLARATION OF CONFLICTS OF INTEREST
The authors, reviewers, editors, and publication staff do not report any relevant conflicts of interest. The opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect those of the Department of Justice.
References
- 1).Relethford J.H. Apportionment of global human genetic diversity based on craniometrics and skin color. Am J Phys Anthropol. 2002. Aug; 118(4): 393–8. PMID: 12124919. 10.1002/ajpa.10079. [DOI] [PubMed] [Google Scholar]
- 2).Sparks C.S., Jantz R.L. A reassessment of human cranial plasticity: Boas revisited. Proc Natl Acad Sci U S A. 2002. Nov 12; 99(23): 14636–9. PMID: 12374854. PMCID: PMC137471. 10.1073/pnas.222389599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Hanihara T., Ishida H. Metric dental variation of major human populations. Am J Phys Anthropol. 2005. Oct; 128(2): 287–98. PMID: 15838862. 10.1002/ajpa.20080. [DOI] [PubMed] [Google Scholar]
- 4).Pilloud M.A., Hefner J.T., Hanihara T., Hayashi A. The use of tooth crown measurements in the assessment of ancestry. J Forensic Sci. 2014. Nov; 59(6): 1493–501. PMID: 25060236. 10.1111/1556-4029.12540. [DOI] [PubMed] [Google Scholar]
- 5).Scott G.R., Turner CG II. The anthropology of modern human teeth: dental morphology and its variation in recent human populations. New York: Cambridge University Press; 1997. 382 p. [Google Scholar]
- 6).Edgar H.J. Prediction of race using characteristics of dental morphology. J Forensic Sci. 2005. Mar; 50(2): 269–73. PMID: 15813536. 10.1520/jfs2004261. [DOI] [PubMed] [Google Scholar]
- 7).Hefner J.T. Cranial nonmetric variation and estimating ancestry. J Forensic Sci. 2009. Sep; 54(5): 985–95. PMID: 19686390. 10.1111/j.1556-4029.2009.01118.x. [DOI] [PubMed] [Google Scholar]
- 8).Hefner J.T., Ousley S.D., Dirkmaat D.C. A companion to forensic anthropology. West Sussex (UK): John Wiley & Sons; c2014. Chapter 14, Morphoscopic traits and the assessment of ancestry; p. 287–310. [Google Scholar]
- 9).Brace C.L. Race is a four-letter word. New York: Oxford University Press; 2005. 326 p. [Google Scholar]
- 10).Hooton E.A. Lecture notes of EA Hooton. Unpublished manuscripts of the Peabody Museum Archives of E.A. Hooton, Cambridge, MA; 1946. [Google Scholar]
- 11).Ousley S., Jantz R., Freid D. Understanding race and human variation: why forensic anthropologists are good at identifying race. Am J Phys Anthropol. 2009. May; 139(1): 68–76. PMID: 19226647. 10.1002/ajpa.21006. [DOI] [PubMed] [Google Scholar]
- 12).Hooton E.A. Lecture notes of EA Hooton. Unpublished manuscripts of the Peabody Museum Archives of E.A. Hooton, Cambridge, MA; 1926. [Google Scholar]
- 13).Brues A.M. Skeletal attribution of race: methods for forensic anthropology. Albuquerque: Maxwell Museum of Anthropology; 1990. Chapter 1, The once and future diagnosis of race; p. 1–9. [Google Scholar]
- 14).Ousley S.D., Hefner J.T. The statistical determination of ancestry. Proceedings of the 57th annual meeting of the American Academy of Forensic Sciences; 2005. Feb 21-26; New Orleans, LA. [Google Scholar]
- 15).Rhine S. Skeletal attribution of race: methods for forensic anthropology. Albuquerque: Maxwell Museum of Anthropology; 1990. Chapter 2, Nonmetric skull racing; p. 10–20. [Google Scholar]
- 16).Hefner J.T. The statistical determination of ancestry using cranial nonmetric traits [Internet]. PhD Dissertation. University of Florida; 2007. [cited 2016 Jun 21]. 126 p. Available from: http://ufdc.ufl.edu/UFE0021200/00001/pdf. [Google Scholar]
- 17).Hefner J.T. Biological affinity in forensic identification of human skeletal remains: beyond black and white. Boca Raton: CRC Press; c2015. Chapter 3, Cranial morphoscopic traits and the assessment of American Black, American White, and Hispanic ancestry; p. 27–41. [Google Scholar]
- 18).Hefner J.T., Ousley S.D. Statistical classification methods for estimating ancestry using morphoscopic traits. J Forensic Sci. 2014. Jul; 59(4): 883–90. PMID: 24646108. 10.1111/1556-4029.12421. [DOI] [PubMed] [Google Scholar]
- 19).Brettell S.A. A validation study examining Hefner's “cranial nonmetric variation and estimating ancestry” [Internet]. University of Tennessee Honors; Thesis Projects; 2013. 25 p. Available from: http://trace.tennessee.edu/cgi/viewcontent.cgi?article=2668&context=utk_chanhonoproj. [Google Scholar]
- 20).Kenyhercz M.W., Klales A.R., Rainwater C.W., Fredette S. The optimized summed scored attributes method for the classification of American Blacks and Whites: a validation study. Proceedings of the 67th Annual Meeting of the American Academy of Forensic Sciences; 2015. Feb 16-21; Orlando, FL. [DOI] [PubMed] [Google Scholar]
- 21).Klales A.R., Kenyhercz M.W. Morphological assessment of ancestry using cranial macromorphoscopics. J Forensic Sci. 2015. Jan; 60(1): 13–20. PMID: 25047253. 10.1111/1556-4029.12563. [DOI] [PubMed] [Google Scholar]
- 22).L'Abbé E.N., Van Rooyen C., Nawrocki S.P., Becker P.J. An evaluation of non-metric cranial traits used to estimate ancestry in a South African sample. Forensic Sci Int. 2011. Jun 15; 209(1-3): 195.e1–7. PMID: 21530118. 10.1016/j.forsciint.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 23).Hurst C.V. Morphoscopic trait expressions used to identify South west Hispanics. J Forensic Sci. 2012. Jul; 57(4): 859–65. PMID: 22372384. 10.1111/j.1556-4029.2012.02080.x. [DOI] [PubMed] [Google Scholar]
- 24).Hefner J.T., Pilloud M.A., Black C.J., Anderson B.E. Morphoscopic trait expression in 'Hispanic' populations. J Forensic Sci. 2015. Sep; 60(5): 1135–9. PMID: 26272587. 10.1111/1556-4029.12826. [DOI] [PubMed] [Google Scholar]
- 25).Ratliff M.D. Evaluating morphoscopic trait frequencies of Southeast Asians and Pacific Islanders [Internet]. Dissertation. University of Montana; 2014. 167 p. Available from: http://scholarworks.umt.edu/cgi/viewcontent.cgi?article=5298&context=etd. [Google Scholar]
- 26).Spradley M.K., Hefner J.T. Using non-metric traits to estimate ancestry in the 21st century. Proceedings of the 85th Annual Meeting of the American Association of Physical Anthropologists; 2016. April 13-16; Atlanta, GA. [Google Scholar]
- 27).Bass W.M. In: Trimble M.K., Editor. Human Osteology: A Laboratory and Field Manual. Columbia (MO): Missouri Archaeological Society; 1995. 361 p. [Google Scholar]
- 28).Hauser G., De Stefano D.F. Epigenetic variants of the human skull. Stuttgart: E. Schweizerbart'sche Verlagsbuchhandlung; 1989. 301 p. [Google Scholar]
- 29).Evteev A.A., Movsesian A.A. Testing the association between human mid-facial morphology and climate using autosomal, mitochondrial, Y chromosomal polymorphisms and cranial non-metrics. Am J Phys Anthropol. 2016. Mar; 159(3): 517–22. PMID: 26567130. 10.1002/ajpa.22894. [DOI] [PubMed] [Google Scholar]
- 30).McDowell J.L., L'Abbé E.N., Kenyhercz M.W. Nasal aperture shape evaluation between black and white South Africans. Forensic Sci Int. 2012. Oct 10; 222(1-3): 397.e1–6. PMID: 22727267. 10.1016/j.forsciint.2012.06.007. [DOI] [PubMed] [Google Scholar]


