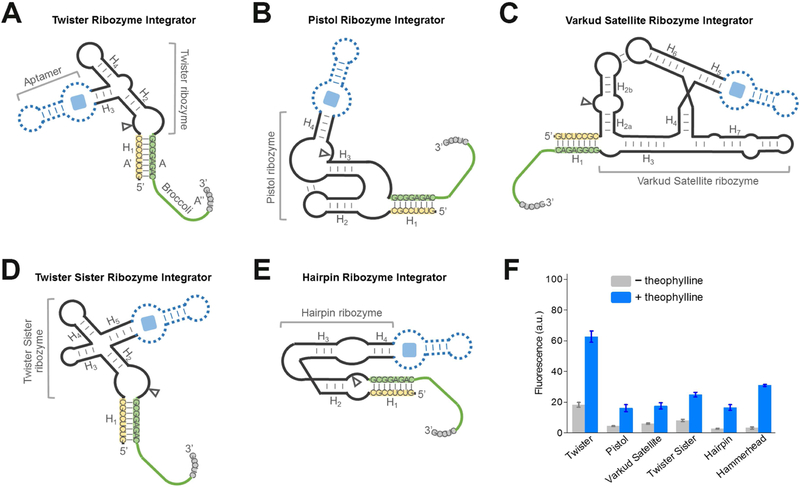
Figure 3. Generation of RNA integrators with diverse natural cis-acting ribozymes.
(A – E) Secondary structure and design of the optimal (A) twister ribozyme integrator, (B) pistol ribozyme integrator, (C) Varkud satellite ribozyme integrator, (D) twister sister ribozyme integrator, and (E) hairpin ribozyme integrator. The integrator is generated by swapping Broccoli-inhibitor A-A’-A’’ complex (green-yellow-grey) into one domain of the ribozyme. Shown are the sequences from the Inhibitor 3 complex. In experiments to test the performance of theophylline-regulated integrator, a theophylline-binding aptamer module (dotted blue) was inserted into the corresponding domain of the ribozyme.
(F) Performance of optimal RNA integrators using each of the six ribozymes. Shown are mean and SEM values of three independent replicates. The fluorescence signal was normalized to that of Broccoli.