TABLE 2.
Glucuronidation kinetic parameters from UGT overexpression in baculosomal cell system
Values represent the parameter estimate (S.E.) by fitting substrate concentration to the simple Michaelis-Menten (MM), Hill, or two-site equation, as described in the Materials and Methods, to metabolite formation velocity using Phoenix WinNonlin (version 7.0).
| Enzyme Source |
Model |
Km,u or S50,u |
Vmax |
n |
Clint,u or Clmax,u |
|---|---|---|---|---|---|
| µM | pmol/min/mg total protein | µl/min/mg protein | |||
| Cabotegravir | |||||
| rUGT1A1 | Two Site | 27 (6)a | 17 (1)a | 0.6 | |
| rUGT1A3 | MM | 46 (7) | 3 (0.1) | 0.06 | |
| rUGT1A7 | MM | 43 (5) | 10 (0.3) | 0.2 | |
| rUGT1A8 | MM | 344 (44) | 40 (2) | 0.1 | |
| rUGT1A9 | Hill | 56 (7) | 61.4 (3) | 1.26 (0) | 1.1 |
| Dolutegravir | |||||
| rUGT1A1 | MM | 216 (26) | 507 (50) | 2.3 | |
| rUGT1A3 | MM | 62 (7) | 18 (0.7) | 0.3 | |
| rUGT1A7 | MM | 9 (2) | 1 (0) | 0.1 | |
| rUGT1A8 | Hill | 44 (5) | 7 (0) | 1.9 (0) | 0.2 |
| rUGT1A9 | Hill | 39 (3) | 39 (2) | 1.9 (0) | 1.0 |
| Raltegravir | |||||
| rUGT1A1 | MM | 260 (17) | 334 (7) | 1.3 | |
| rUGT1A3 | Hill | 41 (2) | 30 (1) | 1.7 (0) | 0.55 |
| rUGT1A7 | MM | 452 (56) | 23 (1) | 0.05 | |
| rUGT1A8 | MM | 386 (57) | 39 (2) | 0.1 | |
| rUGT1A9 | Hill | 193 (12) | 459 (14) | 1.45 (0) | 1.3 |
Clint = Vmax/Km or Clmax = 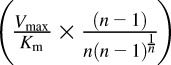 . n, Hill coefficient in the Hill equation.
. n, Hill coefficient in the Hill equation.
The low-affinity enzyme substrate concentration versus velocity did not saturate, and the estimated Km (>2 mM) and Vmax values were unreliable. Thus, only the high Km is presented and calculated for Clint,u.
