Figure 4.
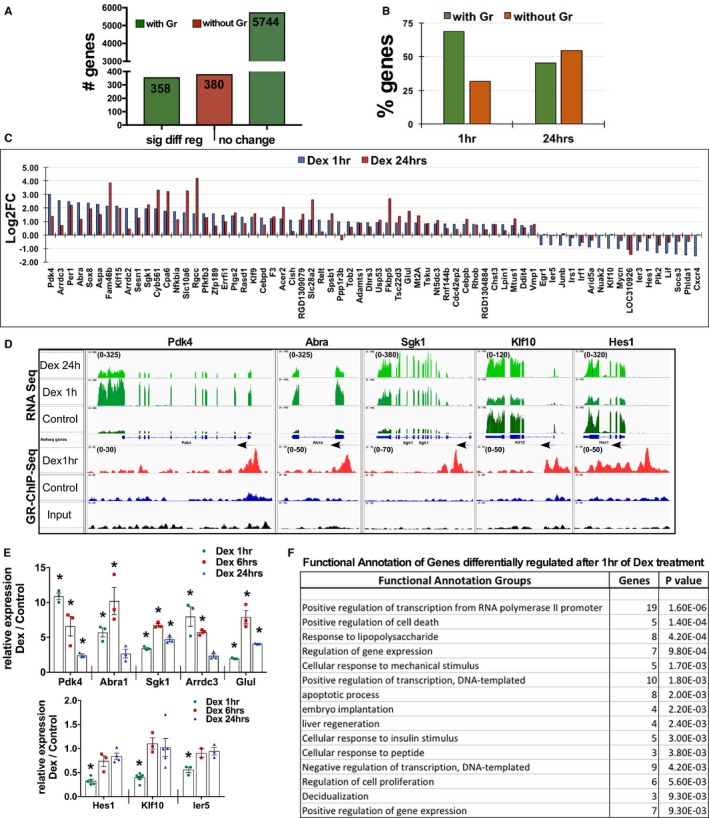
Early targets of GR include regulators of pol II–dependent transcription and gene expression in cardiomyocytes. A, The graph represents the numbers of genes with or without GR association, sorted into genes that show significant differential regulation and genes that do not show significant change in transcript abundance. B, The graph represents the number of genes differentially regulated and the status of GR genomic association at the 1‐hour and 24‐hour time points of dexamethasone treatment. C, Genes with significant change in mRNA transcript that are associated with genomic GR binding at 1 hour along with corresponding change after 24 hours of dexamethasone treatment are graphed and presented based on log2‐fold change (Log2FC) value. D, Integrated genomic viewer screenshots of selected representative genes with integrated RNAseq and GR ChIP‐Seq data, showing change in the transcript after 1 and 24 hours of dexamethasone along with genomic GR binding status of genes that are differentially regulated at 1 hour dexamethasone treatment compared with control (ethanol) cardiomyocytes. Arrows indicate the direction of transcription of that gene, numbers in brackets in the Y axis indicate the values on signal tracks for GR ChIP‐Seq and RNAseq for each gene. The values were kept the same within the samples for each gene. E, Relative transcript abundance of selected genes as measured by quantitative polymerase chain reaction in cardiomyocytes treated with dexamethasone for 1, 6, or 24 hours. Error bars represent SEM, *P<0.05 compared with control, n=3. F, Functional annotation of genes that show significant differential regulation after 1 hour of dexamethasone treatment that are associated with genomic GR, as analyzed using DAVID; top 15 groups are shown with number of genes and P value (full table with all the categorized genes in Figure S4). DAVID indicates Database for Annotation, Visualization and Integrated Discovery; Dex, dexamethasone; GR, glucocorticoid receptor.
