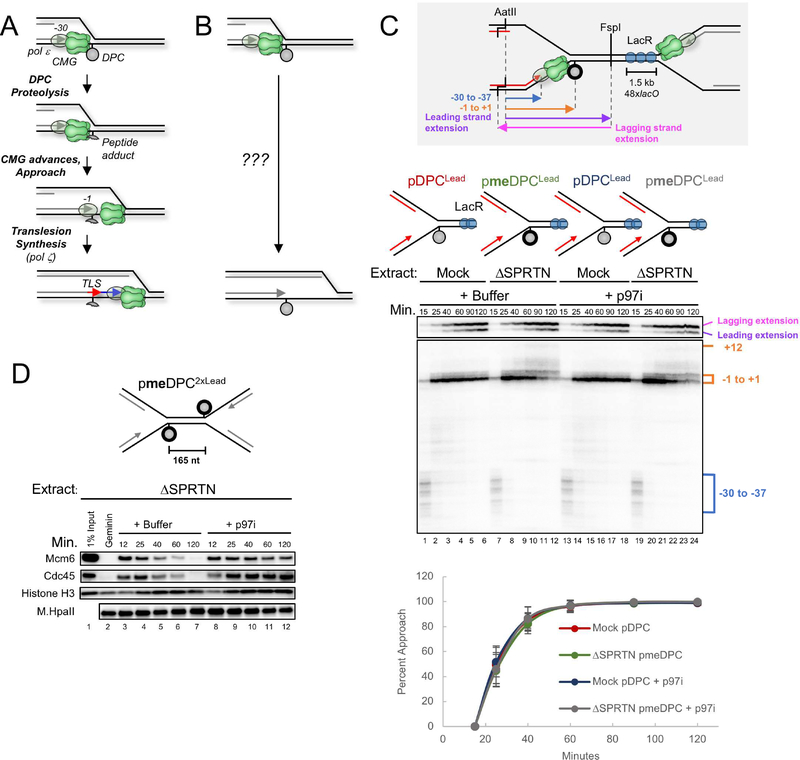
Figure 1. Disappearance of the CMG footprint at DPCLead is unaffected by proteolysis or p97 inhibition.
(A) Previous model of replication-coupled DPC repair (Duxin et al., 2014). (B) Schematic of what happens in the presence of Ub-VS (Duxin et al., 2014). (C) pDPCLead or pmeDPCLead were pre-bound with LacR to prevent one replication fork from reaching the DPC. The plasmids were replicated in mock-depleted or SPRTN-depleted egg extract containing 32P[α]-dATP and supplemented with buffer or the p97 inhibitor NMS-873 (p97i). At different times, DNA was recovered and digested with AatII and FspI, separated on a denaturing polyacrylamide gel, and visualized by autoradiography. Grey inset: Schematic of nascent leading strand products released by AatII and FspI digestion of pmeDPCLead or pDPCLead. The lower autoradiogram shows nascent leading strands generated by the rightward replication fork, and the upper autoradiogram shows both extension products. Blue bracket, CMG footprint (−30 to −37); orange bracket, products stalled at the adducted base (−1 to +1). The percentage of leading strands that approached from the −30 cluster to the −1 cluster was quantified (see methods), and the mean of n=5 experiments is graphed. Error bars represent the standard deviation. See Figure S1E for description of −1 to +12 products in lanes 7–12 and 19–24. (D) pmeDPC2xLead was replicated in SPRTN-depleted egg extracts and supplemented with buffer or p97i. At different times, plasmid-associated proteins were recovered and blotted with the indicated antibodies. Samples were also examined for DPC proteolysis (Figure S1D). A model of CMG unloading from this template is shown in Figure S1G.