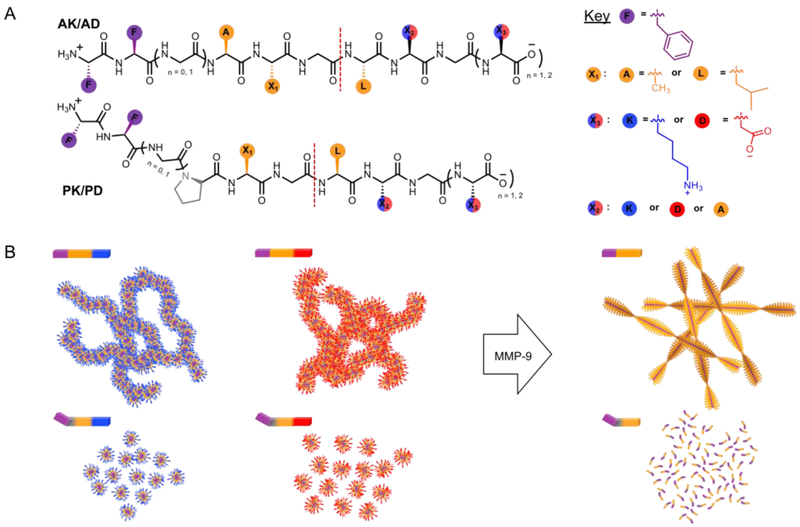
Figure 1.
Sequence dependent peptide nanostructures. (A) Chemical structure and (B) cartoon of self-assembling peptide amphiphiles that respond to MMP-9 action. Positive (blue) or negative (red) charges on the nanoparticle electrostatically recruit or repel MMP-9 to influence enzyme kinetics. Self-assembling (purple) and MMP-9 cleavable segments (gray and/or orange) dictate the susceptibility of the nanostructures to MMP-9 hydrolysis by forming ordered/disordered structures, and control the fiber formation or disassembly of the post-enzymatic products. Red dash line indicates the scissile bond.