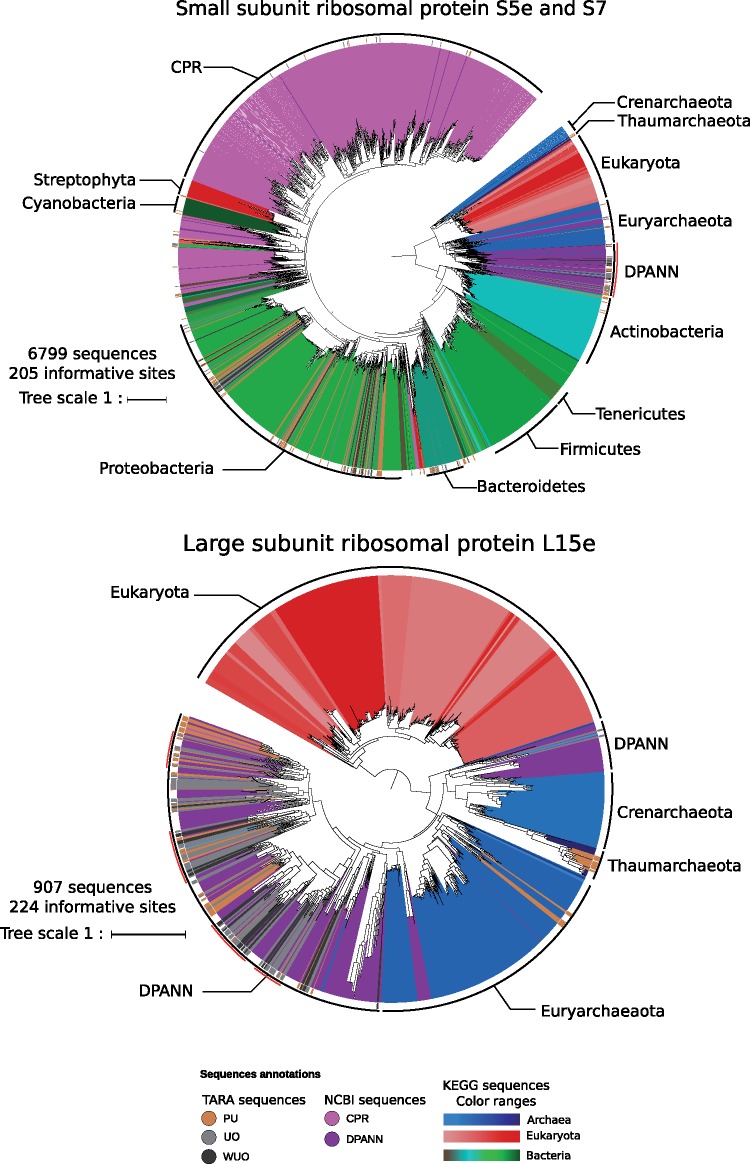
Fig. 3.
—Phylogenetic trees of ribosomal proteins S5e and S7 (top) and L15e (bottom). Sequences were aligned using MAFFT in auto mode and trimmed with TrimAl. Trees were constructed using IQ-TREE with LG + G4 models and ultrafast bootstrap approximation (Minh et al. 2013). The trees were rooted between Archaea and Bacteria and branches with bootstrap values <50% were collapsed. The number of informative sites and branch length scale bars (substitutions per site) are shown. Environmental sequences are highlighted by a colored bar in the outer ring. The sequences were from three sources: 1) environmental sequences from the TARA Oceans data sets; 2) CPR and DPANN sequences from assemblies available in NCBI; and 3) other reference sequence from KEGG. Sequences are colored by taxonomic annotation. Archaeal sequences found in 037, 038, and 039 MES sampling sites are highlighted by a red arc. Abbreviations: PU, Potentially Ultrasmall; UO, Ultrasmall Only; and WUO: Widespread Ultrasmall.