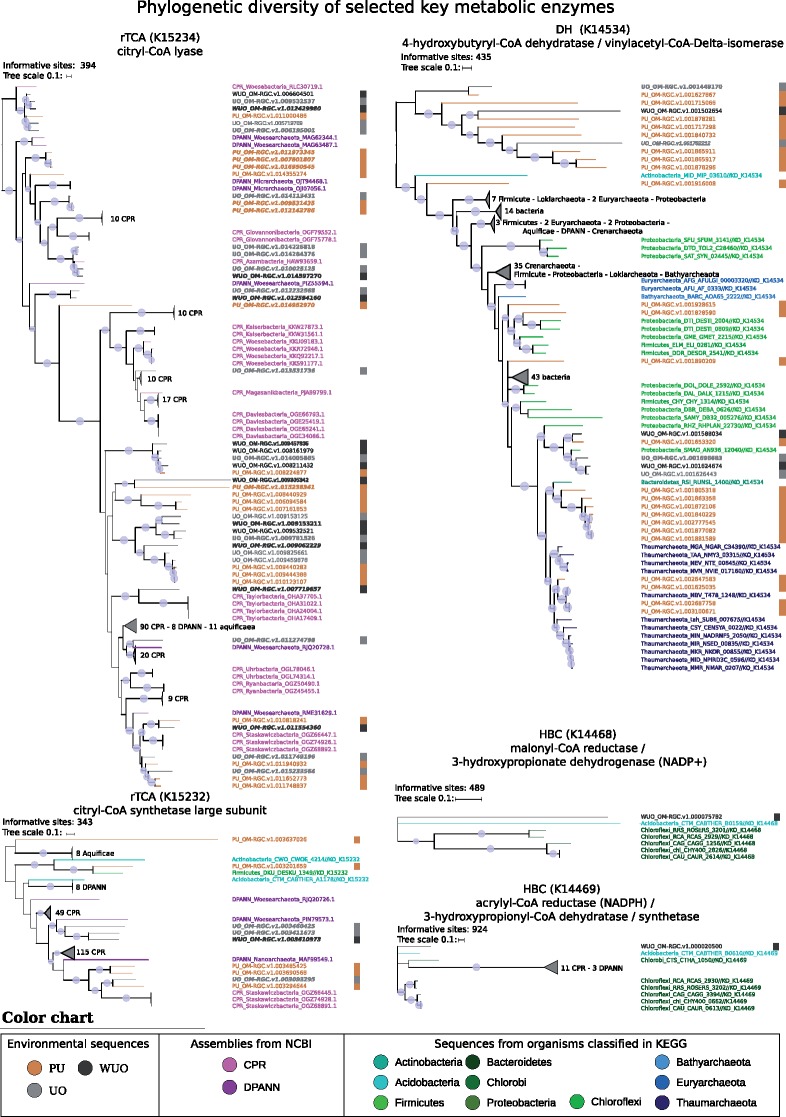
Fig. 4.
—Phylogenetic trees of selected key enzymes in three carbon fixation pathways. Trees were reconstructed using maximum likelihood on trimmed alignments. The number of informative sites and branch length scale bars (substitutions per site) are shown for each tree. Sequences used are from KEGG, NCBI (CPR and DPANN) and from the TARA Oceans data set. Trees were midpoint rooted. Bootstraps were computed using 1,000 iterations of ultrafast bootstrap approximation. Branches with bootstrap <50% were collapsed, a light blue dot highlights branches with bootstrap values >80%. Environmental sequences are highlighted by a colored bar on the right of each tree. Sequence names: environmental sequences were formatted as (PU, UO, WUO)TARA identifier. KEGG sequences were formatted as phylum_KEGG_identifier. NCBI sequences were formatted as (CPR/DPANN)_phylum_proteinID. For readability, some clades were collapsed and are represented by a dark triangle with the description of the clade’s sequences. Abbreviations: rTCA, reductive tricarboxylic acid cycle; DHC: dicarboxylate–hydroxybutyrate cycle; HBC, 3-hydroxypropionate bi-cycle; PU, Potentially Ultrasmall; UO, Ultrasmall Only; and WUO, Widespread Ultrasmall Only.