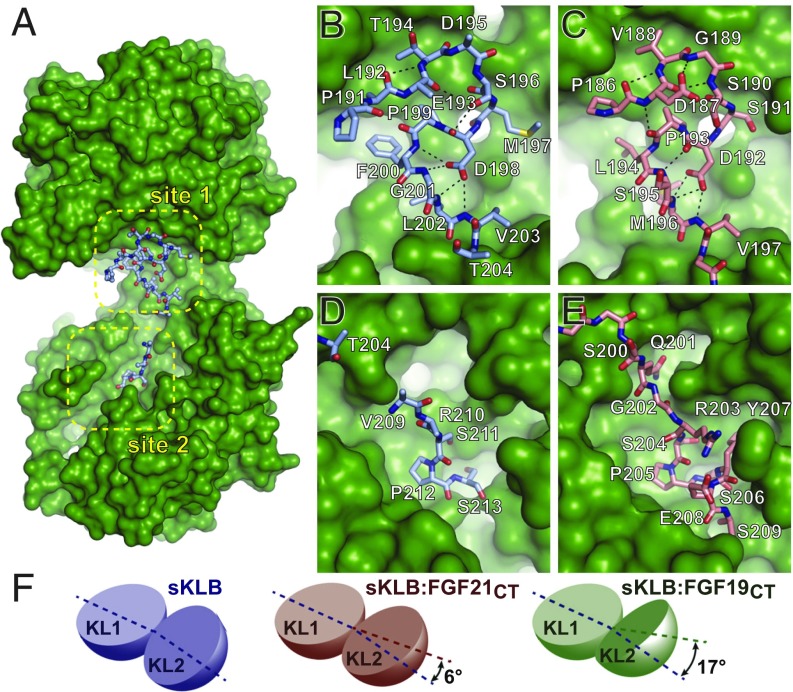
Fig. 2.
Comparison of ligand-binding sites in crystal structures of sKLB-ligand complexes. (A) Surface representation of sKLB (green) highlighting two binding sites, site 1 and site 2, of FGF19CT (light blue, ball-and-stick). Nb30 is omitted for clarity. (B–E) Regions of (B and D) FGF19CT (light blue, stick) and (C and E) FGF21CT (salmon, stick) that interact with (B and C) site 1 of sKLB and (D and E) site 2 of sKLB. Site 1-interacting regions in both (B) FGF19CT and (C) FGF21CT form consecutive turns through intramolecular hydrogen bonds indicated as black dashed lines. S-P-S motifs in site 2-interacting regions in both (D) FGF19CT and (E) FGF21CT interact with a pseudosubstrate-binding pocket in KL2 of sKLB. (F) Diagrams depicting change interdomain angles across various crystal structures of sKLB. Angles are shown as a difference in comparison with the structure of sKLB:Nb914 (PDB ID: 5VAN). Changes in sKLB interdomain angles upon binding FGF19 or FGF21 vary not only in absolute angle values (16.9° and 5.9°, respectively), but also in the direction of movement of KL2 in respect to KL1, which are calculated as different contributions of twist and closure components to the resulting movement. Twist is movement around the axis parallel to the line joining the centers of mass of domains, and closure is movement around the axis perpendicular to this line (21). The contributions from twist and closure components are 41 and 59% for sKLB:FGF19CT and 23 and 77% for sKLB:FGF21CT, respectively.