Figure 1.
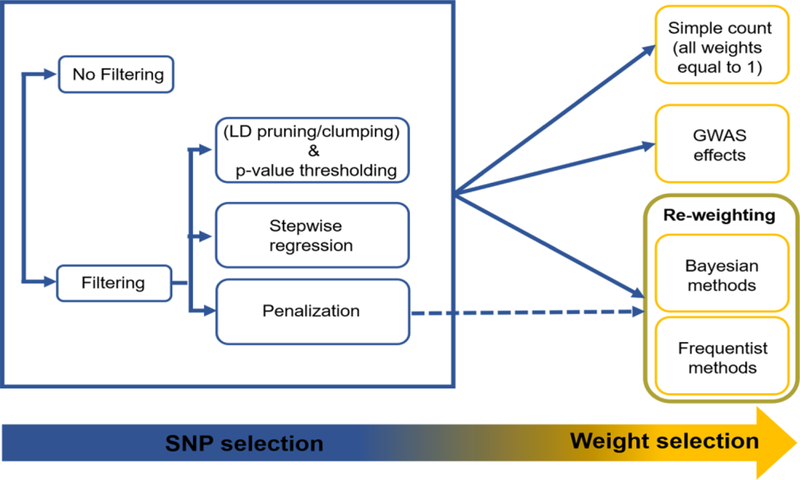
Key Figure. Polygenic risk score calculation. Step1) SNP selection (with or without filtering), Step 2) weight calculation: Candidate SNPs can be assigned a weight of 1 (PRS is a simple sum of SNP alleles) or weighed using existing GWAS-derived effect sizes. Alternatively, one can re-calculate the SNP weights (re-weighting), that is, estimate new weights by including the SNPs in a regression model (e.g., Cox). Penalization techniques (either frequentist e.g., Lasso or Bayesian e.g., LDpred) can also be used for re-weighting. These methods can achieve SNP selection and weight estimation simultaneously, by setting some of the SNP weights to zero. Penalization methods can be either applied on the filtered or on the original SNP list.
