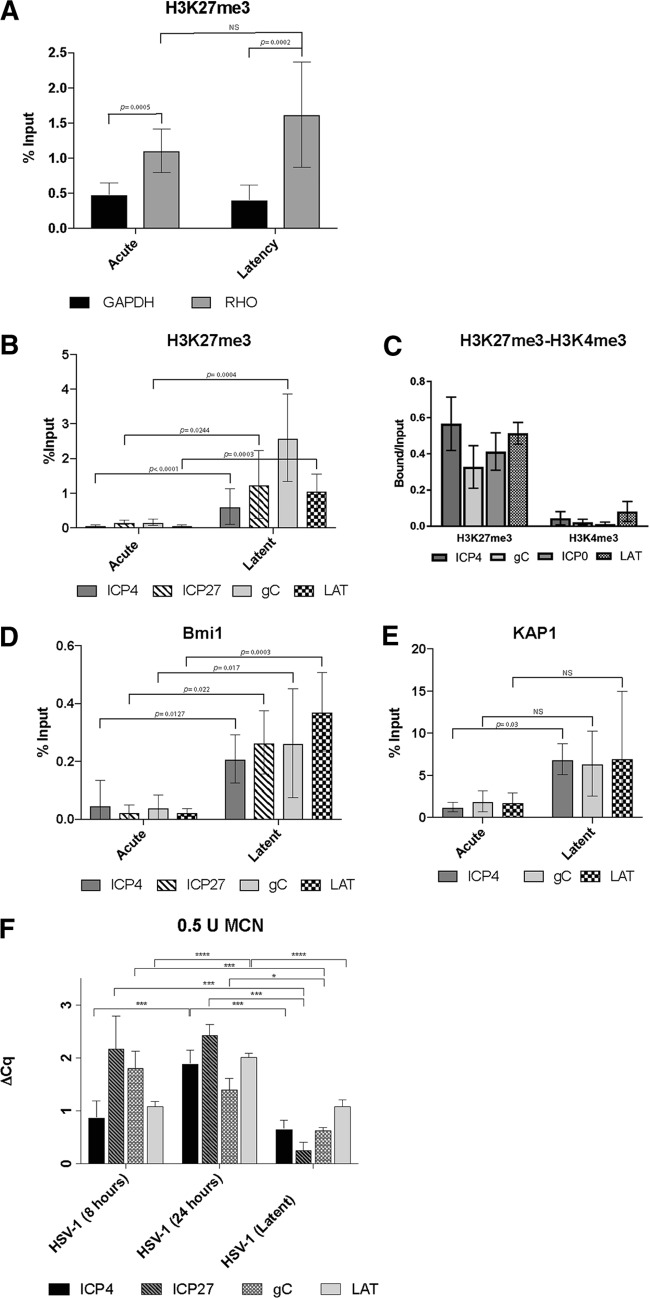
FIG 3.
Chromatin analysis of HSV-1 in acutely and latently infected hiPSC-derived CNS neurons. (A) Validation of the ChIPs using anti-H3K27me3 by analyzing the results of ChIP-qPCR with positive-control PCR primers/probe to rhodopsin (RHO) compared to those of ChIP-qPCR with a negative-control PCR primer/probe to GAPDH. (B to E) Comparison of the enrichment of H3K27me3 (B and C), H3K4me3 (C), the polycomb group protein Bmi1 (D), and the coregulator of the Krüppel-associated box-containing zinc finger proteins (KAP1) (E) at the indicated viral promoter regions in acutely and latently infected hiPSC-derived neuronal cultures via ChIP analysis. The data represent averages from three independent experiments. The ChIP-qPCR data in panels A, B, D, and E were normalized using the percent input method, while in panel C, the relative quantities of enrichment are represented as bound/unbound. Error bars represent standard deviations (SD). P values were determined using Student's t test. (F) Analysis of HSV-1 chromatin accessibility to MCN by ChART-PCR. hiPSC-neurons were acutely infected with an HSV-1 construct expressing reporter genes EGFP and RFP under the control of viral promoters for 8 h and 24 h and latently infected with HSV-1 for 7 days at an MOI of 0.3. Following MCN digestion and DNA extraction, real-time qPCR was conducted using primers specific to promoter regions of viral genes. MCN accessibility was determined by calculating the difference in the amounts of undigested DNA and digested DNA (ΔCq). Error bars represent the standard deviation of the ΔCq values. P values were determined using a post hoc Tukey test. *, P ≤ 0.05; ***, P ≤ 0.001; ****, P < 0.0001; NS, not significant.