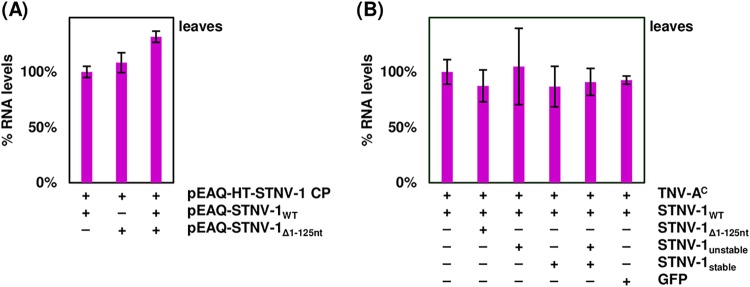
FIG 5.
(A) RT-qPCR analysis of total viral RNAs (HT-STNV-1 CP, STNV-1WT, and STNV-1Δ1-125) in the leaves of plants agroinfiltrated with pEAQ-HT-STNV-1 CP in addition to pEAQ-STNV-1WT, STNV-1Δ1–125, or both, normalized to N. benthamiana 18S rRNA. (B) RT-qPCR analysis of STNV-1WT RNA in the leaves of plants infected with STNV-1 WT and TNV-AC in addition to STNV-1unstable, STNV-1stable, or both, normalized to N. benthamiana 18S rRNA. Samples were collected at 7 days postagroinfiltration or postinfection of N. benthamiana plants. At least three independent repetitions were performed in triplicate, and error bars represent standard deviation. The presence or absence of nonreplicating RNAs can be found in Table 1. At least three independent repetitions were performed in triplicate, and error bars represent standard deviation.