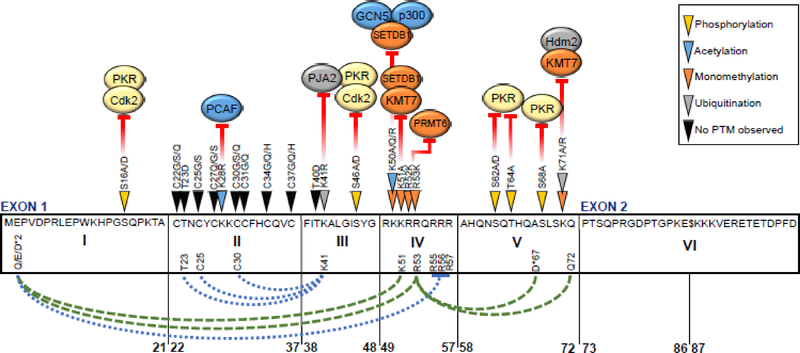
Fig. 2. Effect of HIV-1 Tat amino acid variation on TAR-dependent LTR transactivation, post-translational modifications, and intramolecular interactions.
Variation within the amino acid sequence of HIV-1 Tat contributes to altered LTR transactivation capacity, modeled on the subtype B HXB2 reference sequence. An alternative STOP codon at position 87 is represented by the symbol “$”. Filled arrows along the length of Tat indicate residues with variation that affects LTR transactivation directly or impairs interactions with, or addition of, any of a number of post-translational modifications (PTM) by corresponding host factors. Kinases CDK2 and PKR shown in yellow; histone acetyltransferases PCAF, GCN5, and p300 in blue; monomethyltransferases KMT7, PRMT6, and SETDB1 shown in orange; and E3 ubiquitin ligases PJA2 and Hdm2 shown in gray. Intramolecular interactions observed between Tat residues (bottom) are shown for subtype B (blue dotted) and subtype D (green dashed) Tat. An asterisk next to an amino acid indicates that the dominant amino acid observed differs from the HXB2 sequence.