Figure 1. Outline of trio binning and haplotype assembly.
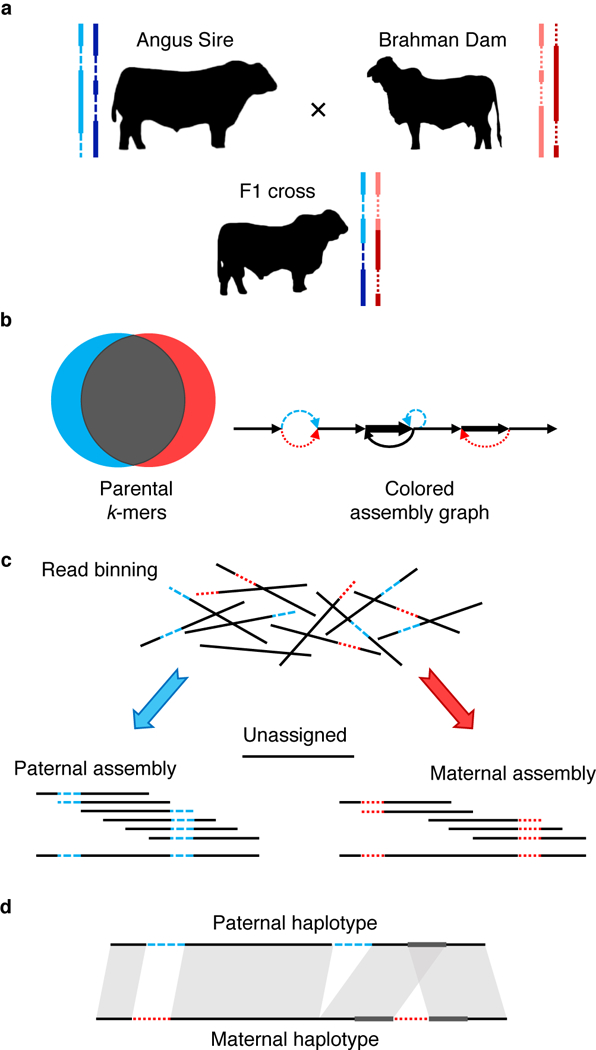
a) Two parents constitute four haplotypes including shared sequence in both parents (solid lines) and sequence unique to one parent (dashed lines). The offspring inherits a recombined haplotype from each parent (blue, paternal; red, maternal). b) Short-read sequencing of the parents identifies unique length-k subsequences (k-mers), which can be used to infer the origin of heterozygous alleles in the offspring’s diploid genome. c) Trio binning simplifies assembly by first partitioning long reads from the offspring into paternal and maternal sets based on these k-mers. Each haplotype is then assembled separately without the interference of heterozygous variants. Unassignable reads are homozygous and can be assigned to both sets or assembled separately. d) The resulting assemblies represent genome-scale haplotypes, and accurately recover both point and structural variation.
