Figure 2. Effect of data characteristics on trio binning.
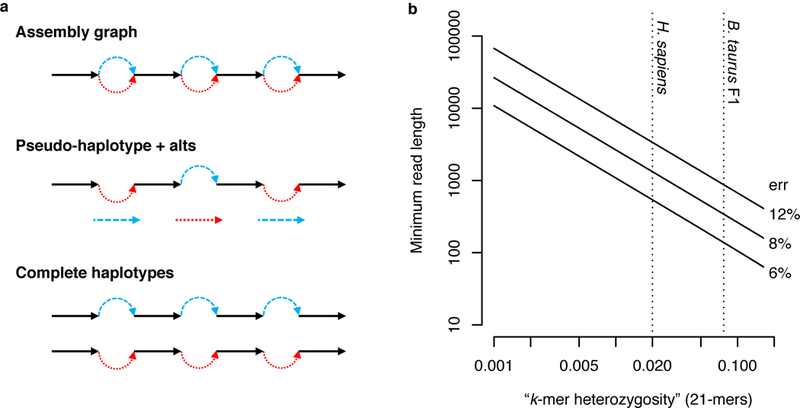
a) Diploid assembly representations shown with homozygous alleles in black and heterozygous alleles (called “bubbles”) colored by haplotype. Graphical representations typically collapse homozygous alleles into a single sequence. A pseudo-haplotype is a path through the diploid graph that separates heterozygous alleles but does not preserve phase between loci. Complete haplotypes represent all alleles and preserve phase across the entire genome. Ability to assign sequencing reads to a haplotype depends on the zygosity of the genome, the sequencing read length, and the sequencing error rate. b) Log-log plot of minimum required read length (y-axis) such that there is a 99% probability of observing at least one haplotype-specific 21-mer per read (negative binomial distribution, Methods), dependent on the sequencing error rate (labels) and fraction of haplotype-specific 21-mers in the genome (x-axis). Dotted vertical lines mark the fraction of heterozygous 21-mers for H. sapiens and the B. taurus F1 cross.
