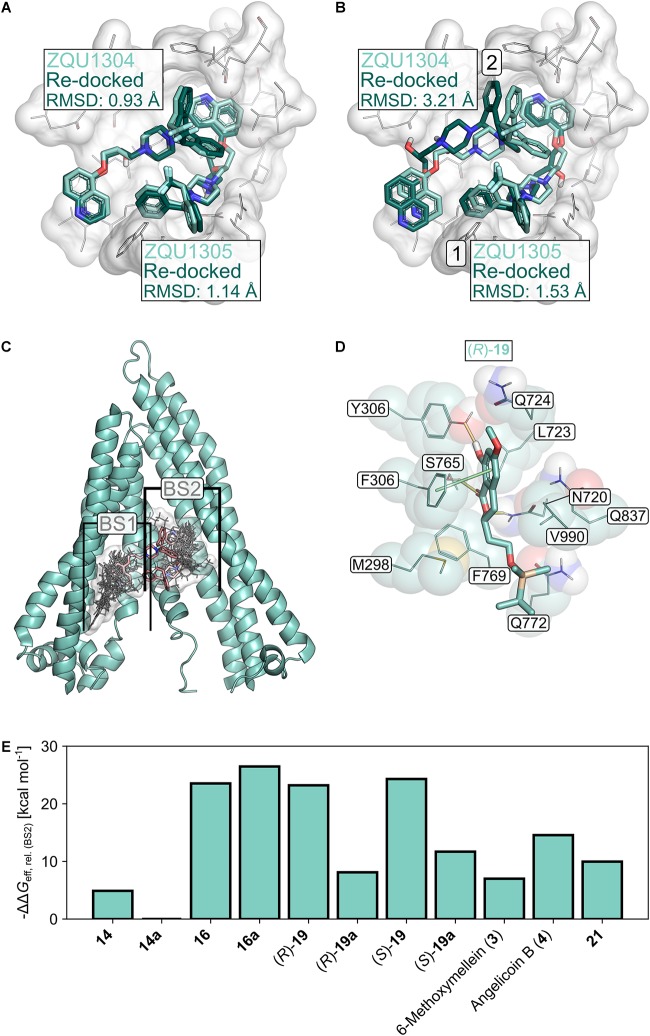
FIGURE 8.
Molecular docking studies. (A) Re-docking of Zosuquidar (ZQU, darker turquoise, sticks representation) into the cryo-EM structure (PDB ID: 6FN1; Alam et al., 2018) of P-gp (white, sticks and cartoon representation, only part of the protein is shown for clarity) while the coordinates of the respective second ZQU molecule (lighter turquoise) were retained from the cryo-EM structure. (B) Sequential re-docking of ZQU into the cryo-EM structure of P-gp. The result of the first and second runs are indicated with a 1 and 2, respectively. Colors and representations are as in (A). (C) Binding sites (denoted as BS1 and BS2) for 3,4-dihydroisocoumarins found during docking. The molecular structures of the 3,4-dihydroisocoumarins are displayed as lines, and their joint molecular surface is rendered in white. Zosuquidar is shown as red sticks. The relevant region in the transmembrane domain of P-gp is shown in cartoon representation (D) Binding mode of (R)-19 in BS2 of P-gp. Hydrogen bonds are displayed as yellow lines, π-stacking interactions as green lines. (E) Relative effective binding energies for 3,4-dihydroisocoumarins in P-gp, calculated by the MM-GBSA approach. To allow for a better comparison with the P-gp transport assay, the energies are expressed as -ΔΔGeff.