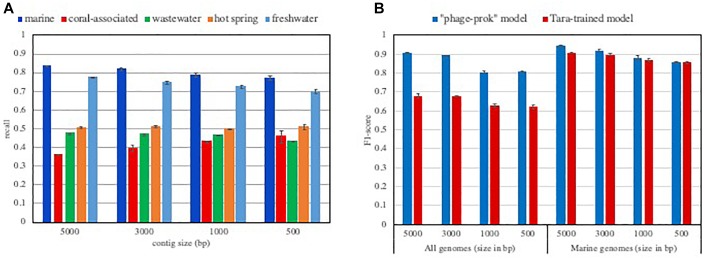
FIGURE 1.
Influence of the training set composition on the model performances. (A) Recall of VirFinder “phage-prok” model on viral contigs isolated in various aquatic ecosystems. The recall was assessed for VirFinder “phage-prok” model when considering viral contigs isolated in various aquatic ecosystems (pelagic, freshwater, hot spring, coral-associated and wastewater). The sequences were downloaded from the IMG V/R env database (methods described in Supplementary File S1 and list of metagenomes used in Supplementary File S2). The viral sequences were broken down to 5000, 3000, 1000, and 500 bp and used to evaluate VirFinder “phage-prok” model. The mean of the recall was calculated for three evaluation sets of 2000 viral sequences each with the exception of the coral-associated evaluation sets composed of 200 viral examples due to the low amount of sequence available for this ecosystem. The error bars correspond to the standard deviation on the three measures. (B) F1-score of classifiers trained on Tara Oceans Metagenomes. Tara-trained models were trained on 10 000 viral and 10 000 prokaryotic sequences from Tara Oceans metagenomes and viromes broken down to 5000 bp. Previous cleaning steps were performed to ensure a low contamination content of the training set (see Supplementary File S1). The F1-score of a Tara-trained model and of VirFinder’s “phage-prok” model was calculated for evaluation sets composed of viruses and prokaryotes isolated in a marine ecosystem (“marine genomes”) or an evaluation set composed of viral and prokaryotic genomes regardless of their origin (“all genomes”). For the “marine evaluation set,” genomes from phages and prokaryotes isolated in marine ecosystems were downloaded from Genbank and the Patric database, respectively, and the sequences were broken down to 5000, 3000, 1000, and 500 bp (see methods in Supplementary File S1 and list of genomes available in Supplementary File S2). The “all genomes” evaluation set is composed of genomes from phages and prokaryotes from RefSeq database published after 2014 (see methods in Supplementary File S1 and list of genomes available in Supplementary File S2). The mean of the F1-score was calculated for three evaluation sets composed of 2000 viral sequences and 2000 prokaryotic sequences. The error bars correspond to the standard deviation on the three measures.