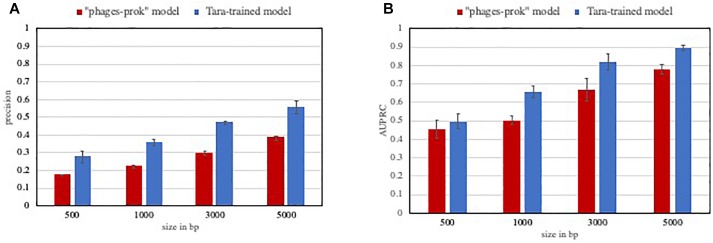
FIGURE 2.
Performance of classifiers on a low viral content evaluation set. The precision (A) and Area under the precision-recall curve (AUPRC) (B) was calculated for VirFinder “phages-prok” model and a Tara-trained model on an imbalanced marine evaluation set. The evaluation set is composed of sequences from genomes from phages and prokaryotes isolated in marine ecosystems, downloaded from Genbank and the Patric database, respectively, and the sequences were broken down to 5000, 3000, 1000, and 500 bp (see methods in Supplementary File S1 and list of genomes available in Supplementary File S2). The mean precision (A) and AUPRC (B) was obtained on three evaluation sets composed of 100 viral sequences and 1900 non-viral sequences. The error bars correspond to the standard deviation on these three measures.