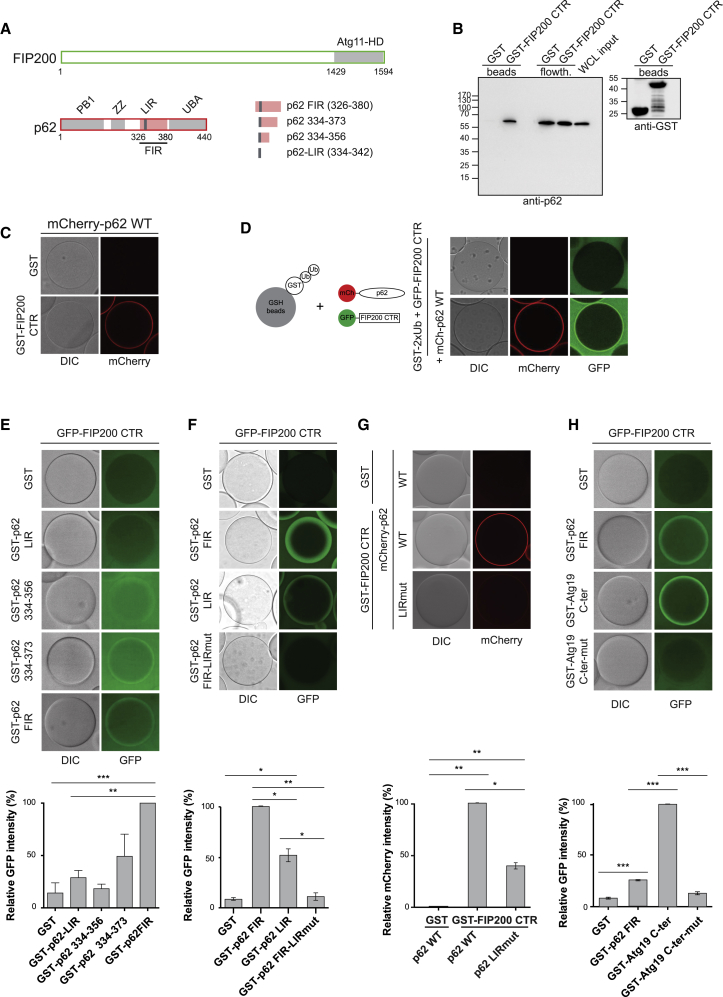
Figure 1.
The FIP200 C-Terminal Region Directly Interacts with p62 in a LIR-Dependent Manner
(A) Schematic representation of FIP200 and p62. The Atg11 homology domain of FIP200 is depicted in gray. The FIP200-interacting region (FIR) of p62 is depicted in pink and the LIR motif in dark gray. p62 constructs covering parts of the FIR are shown on the right.
(B) GST or GST-FIP200 CTR were coupled to glutathione (GSH) beads and incubated with HeLa cells lysates (200 μg). Beads were washed, and the beads/flow-through fractions were analyzed by western blot using anti-p62 antibody. The bound sample was probed with anti-GST to visualize the amount of bait protein on the beads.
(C) GSH beads were coated with GST or GST-FIP200 CTR and incubated with recombinant mCherry-p62 (2 μM). Beads were imaged by microscopy.
(D) The experimental setup is shown on the left. GSH beads were coated with GST-2x ubiquitin. Excess GST-2x ubiquitin was washed off, and beads were incubated with mCherry-p62 (2 μM) and GFP-FIP200 CTR aa 1429–2594 (5 μM). After 1 h incubation, beads were imaged by microscopy.
(E) GSH beads were coated with the indicated GST-p62 constructs and incubated with GFP-FIP200 CTR aa 1429–1594 (5 μM). Beads at equilibrium were imaged by microscopy. Protein inputs are shown in Figure S1B. The GFP signal on the beads was normalized to the signal of GFP-FIP200 CTR bound to GST-p62 FIR-coated beads. Average intensity and SEM for n = 3 are plotted. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(F) Same experimental setup and quantification used in (E). Beads were imaged by microscopy. Protein inputs are shown in Figure S1C. The GST-p62 FIR-LIRmut construct carries a mutation in the LIR motif (335DDDW338 > AAAA).
(G) Same experimental setup used in (C). In the mCherry p62 LIRmut residues 335-338 are mutated to Ala. Protein inputs are shown in Figure S1D. The mCherry signal on the beads was normalized to the signal of mCherry-p62 WT on GST-FIP200 CTR-coated beads. The average intensity and SEM for n = 3 are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(H) The same experimental setup used in (E) and (F) was used to test the interaction of the Atg19 C terminus (Atg19 C-ter) with the FIP200 CTR. In the Atg19 C-ter-mut, the three LIR motifs (Figure S1F) are mutated. The graph on the bottom shows the average GFP intensity, normalized to the signal of GFP-FIP200 CTR binding to GST-Atg19 C-ter, and SEM for n = 3. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001. Protein inputs are shown in Figure S1E.
See also Figure S1.