Figure 5.
p62 LIR Motif Binding Depends on a Positively Charged Pocket in FIP200 CTR
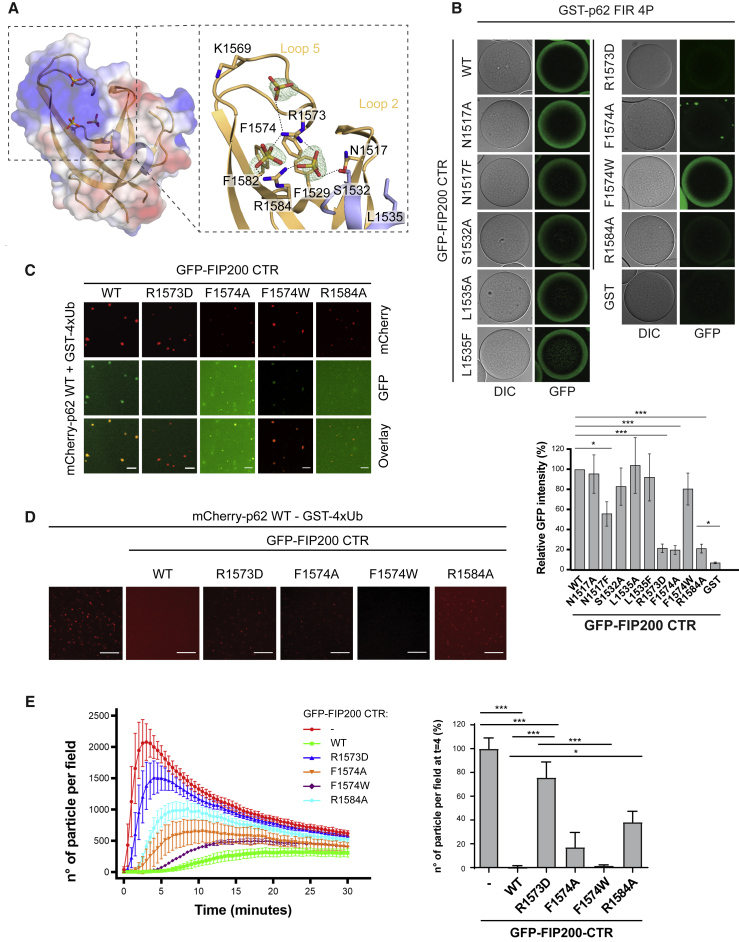
(A) Electrostatic surface potential of the FIP200 Claw domain. Positively and negatively charged surfaces are colored in blue and red, respectively. The coordination of sulfate ions and amino acids of interest are shown as sticks.
(B) GSH beads were coated with GST-p62 FIR 4P, incubated with the indicated GFP-FIP200 CTR (aa 1458–1594) mutants and imaged by microscopy. For each sample the GFP intensity was normalized to the signal of GFP-FIP200 CTR WT on GST-p62 FIR 4P-coated beads. Average intensity and SEM for n = 3 are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001. Protein inputs are shown in Figure S5C.
(C) mCherry-p62 (2 μM) was incubated with GST-4x ubiquitin (10 μM) to form condensates in solution. Pre-formed condensates were incubated with 1 μM GFP-FIP200 CTR (aa 1458–1594). The recruitment of GFP-FIP200 CTR to p62-ubiquitin clusters was monitored by confocal microscopy. Scale bar, 5 μm.
(D) mCherry-p62 was incubated with GST-4x ubiquitin in the presence or absence of the GFP-FIP200 CTR (WT or mutants). The formation of p62-ubiquitin condensates was monitored over 30 min. Images show p62-ubiquitin clusters at t = 4 min. Scale bar, 25 μm. Protein inputs are shown in Figure S5E.
(E) Quantifications of the experiment in (D). For each sample, the number of particles per field is plotted against time (left). Number of particles per field at t = 4 min is plotted on the right. For each sample, the particle number was normalized to the average number of p62-ubiquitin clusters formed in absence of FIP200 CTR. Averages and SEM for n ≥ 3 are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
See also Figure S5.