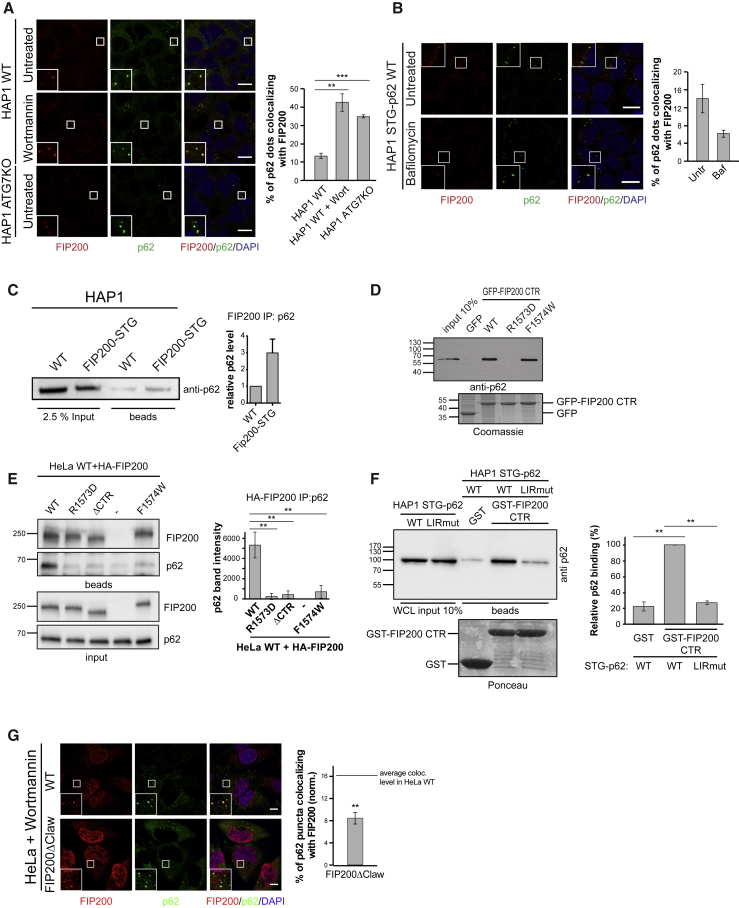
Figure 6.
p62-FIP200 Interaction in Cells
(A) Colocalization analysis of p62 and FIP200 in HAP1 cells (WT or ATG7KO) left untreated or treated with wortmannin (1 μM) for 1 h. Endogenous p62 and FIP200 were detected by immunofluorescence. Scale bar, 10 μm. Average percentages of colocalization and SEM for n = 3 are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(B) Immunofluorescence of p62 and FIP200 in HAP1 STG-p62 cell line left untreated or treated with bafilomycin (400 nM) for 1 h. p62 was detected through the GFP tag fused to the endogenous protein and FIP200 was detected by immunofluorescence. Scale bar, 10 μm. Average percentages of colocalization and SEM (n = 2) are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(C) The C terminus of endogenous FIP200 was tagged in HAP1 cells with GFP-TEV-Strep (FIP200-STG). Affinity purification was performed using HAP1 WT or FIP200-STG cells and the bound material was analyzed by western blotting with anti-p62. The intensities of the p62 bands were normalized for the total level of p62 in the lysate (input). Average p62 levels and SD for n = 4 are shown. Three additional replicates of the immunoprecipitation are shown in Figure S6C.
(D) GFP-Trap beads coated with GFP or GFP-FIP200 CTR (aa 1458–1594) variants were incubated with cell lysates and analyzed by western blotting. Loading control of the bait proteins is shown below the blot.
(E) Anti-HA co-immunoprecipitation in HeLa cells transfected with HA-FIP200. The intensities of the p62 bands were measured and normalized to the amount of the respective bait (HA-FIP200). Average band intensities and SEM (n = 3) are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(F) Pull-down of p62 WT and LIRmut from HAP1 STG-p62 cell lysates was performed as in Figure 1B using GST/GST-FIP200 CTR as bait. Loading control of the bait proteins is shown below the blot. Band intensities were measured and normalized to the intensity of p62 WT binding to FIP200 CTR WT. Average band intensities and SEM (n = 3) are shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
(G) The Claw domain of endogenous FIP200 was deleted in HeLa cells by CRISPR/Cas9 (HeLa FIP200ΔClaw). Cells were treated with wortmannin (1 μM) for 3 h, and p62 and FIP200 were detected by immunofluorescence. Scale bar, 10 μm. The percentage of p62 puncta colocalizing with FIP200 in FIP200ΔClaw cells is compared to the average level of colocalization in WT cells. Average colocalization and SEM for n = 3 is shown. Significant differences are indicated with ∗ when p value ≤ 0.05, ∗∗ when p value ≤ 0.01, and ∗∗∗ when p value ≤ 0.001.
See also Figure S6.