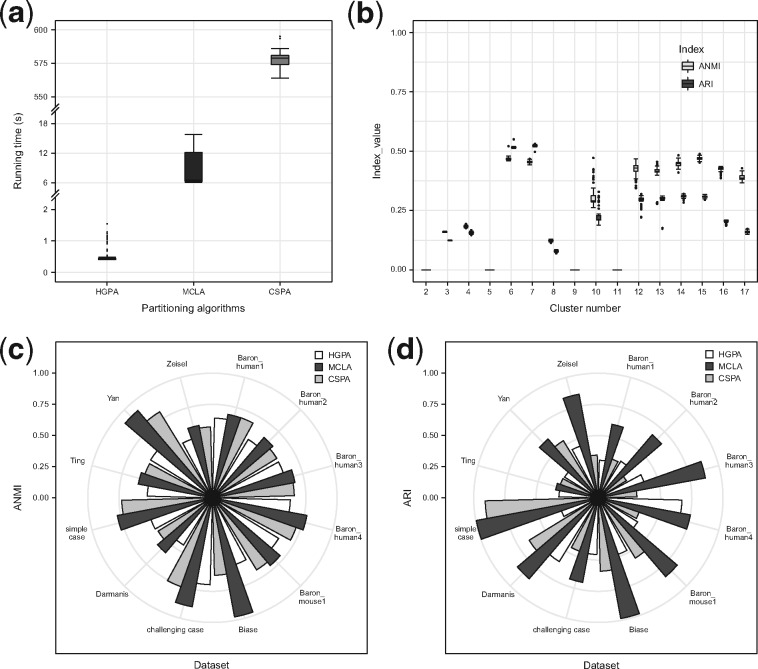
Fig. 5.
Benchmark of the three hypergraph partitioning algorithms: HGPA, MCLA and CSPA. (a) Running time for 3-way partitioning of simple case PBMC mixture dataset with 28 733 single cells using each of the three partitioning algorithms. Each algorithm was applied 100 times. (b) Stability of HGPA from 100 runs using simple case PBMC mixture dataset with 28 733 single cells. (c) Similarity between consensus clustering and individual solutions in 12 benchmarking datasets, measured by average normalized mutual information (ANMI). (d) Performance of the three partitioning algorithms, measured by ARI, across the 12 benchmarking datasets