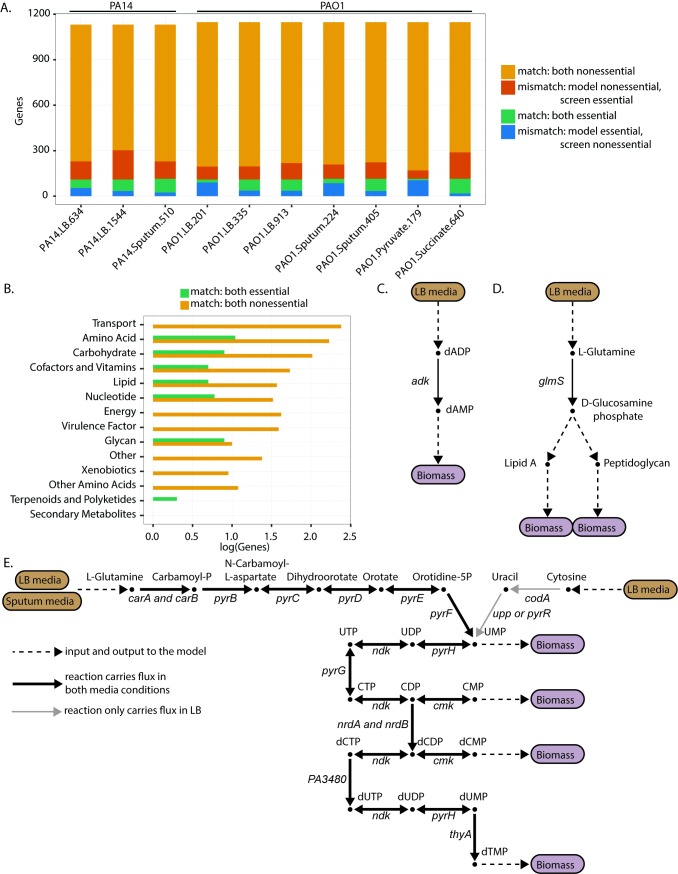
Fig 4. Contextualization of gene essentiality datasets using genome-scale metabolic network reconstructions.
(A). Comparison of model essentiality predictions to in vitro essentiality screens. In silico gene knockouts were performed for both PA14 and PAO1 genome-scale metabolic network reconstructions to predict essential genes. Model-predicted essential genes were compared to the candidate essential genes for each in vitro screen. The bars show the result of this comparison, with orange indicating the number of genes for which both the model and experimental screen identified the gene as nonessential (match: both nonessential), red indicating the number of genes for which the model identified the gene as nonessential whereas the screen identified the gene as essential (mismatch: model-nonessential, screen-essential), green indicating the number of genes for which both the model and experimental screen identified the gene as essential (match: essential), and blue indicating the number of genes for which the model identified the gene as essential whereas the screen identified the gene as nonessential (mismatch: model-essential, screen-nonessential). (B). Functional subsystems for PA14 consensus essential and nonessential genes that were also correctly predicted to be essential or nonessential in the PA14 GENRE. Consensus essential and nonessential genes were identified for PA14 by comparing all three LB screens and determining genes essential or nonessential in all three screens. (C and D). Metabolic pathways demonstrating essentiality for the consensus essential genes adk and glmS, respectively. Dashed lines represent inputs and outputs of the pathway, or, as in D, multiple steps. Brown boxes indicate media inputs, while purple boxes indicate biomass outputs. Metabolites are labeled beside the nodes, with bold metabolites indicating biomass components. Genes associated with the specific reaction are indicated. (E). Flux activity in pyrimidine metabolism under both sputum and LB media conditions. Consensus LB essential genes were compared to consensus sputum essential genes for PAO1. The PAO1 GENRE was used to explain differences in essentiality between the two media-types. Black lines indicate that the reaction is capable of carrying flux under both sputum and LB conditions, while the gray lines indicate that the reaction does not carry flux in sputum conditions but does in LB conditions. Brown boxes are media inputs, purple boxes are biomass outputs. Metabolites are labeled above the nodes, with bold metabolites indicating biomass components. Many of these metabolites are involved in many reactions beyond pyrimidine metabolism. Gene-protein-reaction relationships are indicated in italics beside each reaction edge.