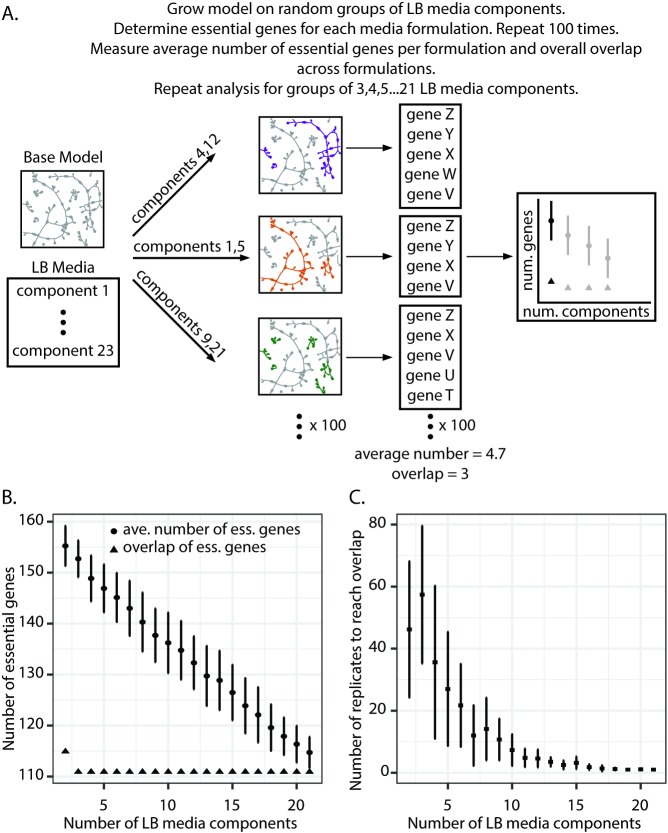
Fig 6. Computational assessment of the impact of LB media composition on condition-independent essentiality.
(A). Pipeline for computational assessment of the impact of LB media formulation on condition-independent essentiality. The PA14 model is grown on different media formulations consisting of random groups of LB components. For instance, two random LB components are selected out of a pool of 23 LB components. The model is grown on these randomly selected pairs and the essential genes for growth on this media formulation are identified. This analysis is repeated 100 times for 100 pairs of LB media components. The average number of essential genes for growth on these random pairs across 100 different formulations is calculated as well as the standard deviation. Additionally, the essential genes common to all 100 different formulations is determined. Ultimately, this random selection of groups of LB media components to support growth of the model and essential gene identification is repeated for groups of three LB components, groups of four, and so on, to groups of 21 LB media components. (B) Impact of LB media formulation on the identification of condition-independent essential genes. Circles represent the average number of essential genes identified in different LB media formulations across 100 comparisons. Triangles represent the shared essential genes (i.e., the overlap) across all 100 comparisons. Error bars indicate standard deviation. (C) Number of replicates needed to converge on shared essential genes in different LB formulations. The pipeline outlined in Panel A was repeated 10 independent times, with 100 replicates per set size. For each iteration, the number of replicates needed to recapture the 111 overlapping genes was calculated. Each data point represents the average number of replicates from the 10 runs. Error bars indicate standard deviation.