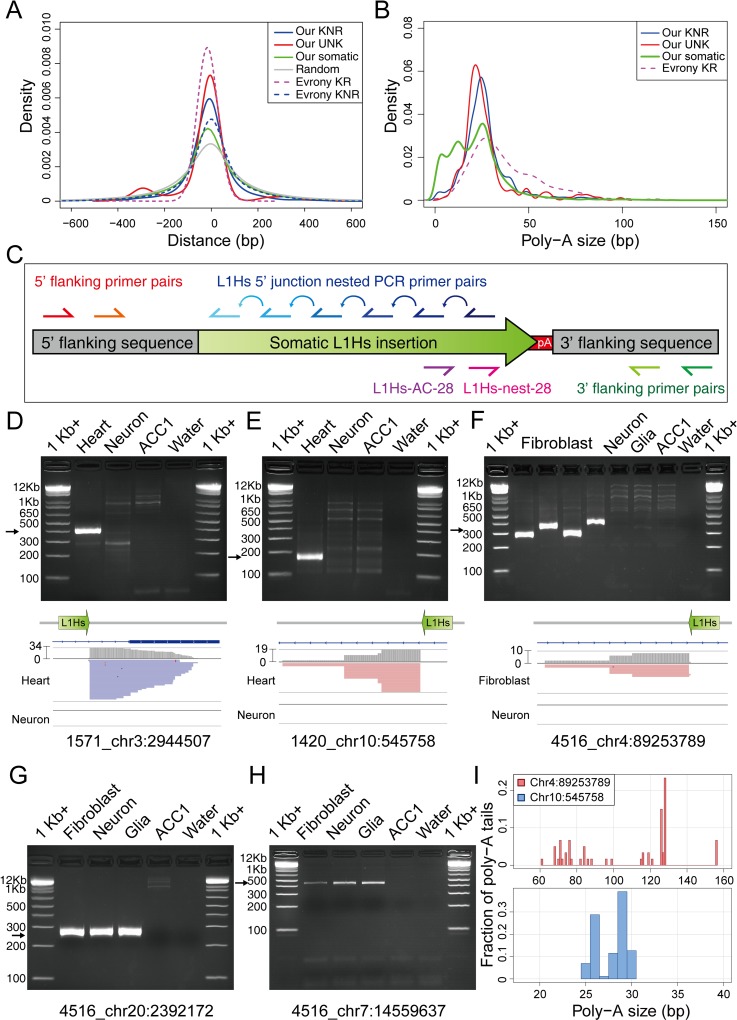
Fig 3. Profiling of somatic L1Hs insertions in multiple human tissues.
(A) The density distributions of L1 EN motifs around L1Hs integration sites. L1 EN motifs included seven specific motifs (TTAAAA, TTAAGA, TTAGAA, TTGAAA, TTAAAG, CTAAAA, TCAAAA). “Evrony KR” and “Evrony KNR” are germline L1Hs insertions identified in Evrony et al. 2012. (B) The density distributions of poly-A tail length for each category of L1Hs insertion. (C) The PCR validation scheme and locations of primers used. (D)–(H) Representative gel images of 3’ nested PCR validation for putative clonal somatic insertions. The Integrative Genomics Viewer screenshots for (D)–(F) showed the coverage track (gray) and the alignment track (blue for read strand [–]; red for read strand [+]) from HAT-seq data. Black arrows indicated bands with target size. 1Kb +: 1 Kb Plus DNA ladder. (I) Polymorphic poly-A tail sizes of clonal somatic insertions measured by capillary electrophoresis. Top: fibroblast-specific somatic L1Hs insertion at chr4:89253789 from Rett patient UMB#4516. Bottom: heart-specific somatic L1Hs insertion at chr10:545758 from Rett patient UMB#1420.