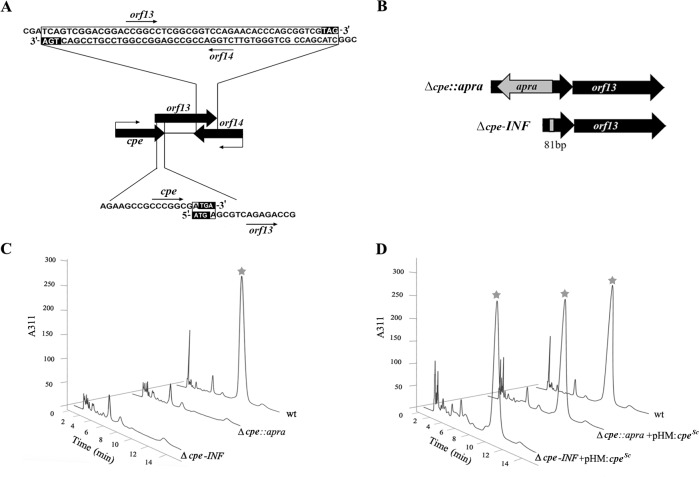
Fig 2. Preparation and analysis of S. clavuligerus cpe deletion mutants.
(A and B) The thick arrows depict genes with arrowheads indicating the direction of transcription. (A) The relative arrangement of genes from the chromosomal locus surrounding cpe in S. clavuligerus is shown, and the bent arrows represent the known promoters for the different transcriptional units. The DNA sequences of the overlapping regions between cpe-orf13 (bottom) and orf13-orf14 (top) are indicated in open boxes, whereas the respective start and stop codons are shown in filled boxes. (B) Diagrammatic representation of the Δcpe::apra and Δcpe-INF mutants, which were prepared such that the 5' and 3' ends of cpe were retained and intervening DNA sequences were replaced by an apramycin resistance cassette or an in-frame 81-bp sequence in the respective mutants. (C and D) HPLC analysis of 96 hour wt S. clavuligerus and different cpe mutant SA culture supernatants for assessing clavulanic acid production using the phosphate buffer system [32]. Peaks corresponding to imidazole-derivatized clavulanic acid are indicated by the star symbol, which were detected at 311nm.