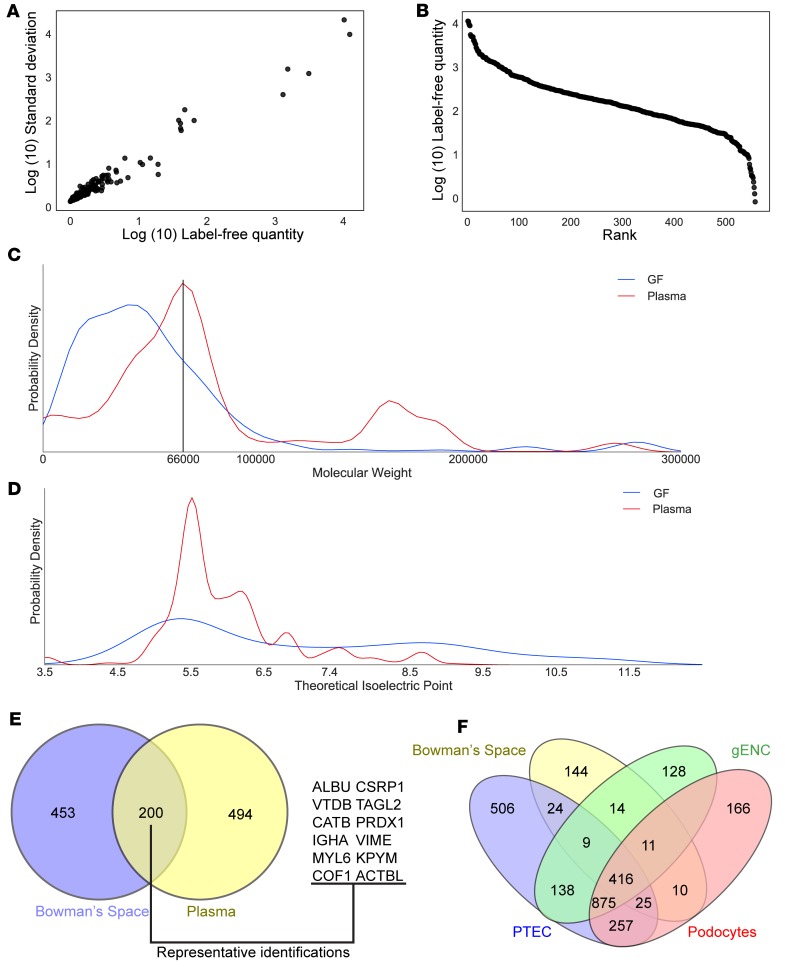
Figure 2. Bowman’s space aspirate contains glomerular filtrate.
(A) Heterogeneity of spectral counts of peptides from Bowman’s space aspiration samples. For each unique protein identification, average spectral count versus standard deviation across 8 samples is displayed. (B) Three-log dynamic range obtained in Bowman’s space aspirate samples using MaxQuant Label-Free Quantification–based (LFQ-based) abundance measurements. For each identification, rank in the total number of identifications is displayed versus log abundance. (C) Kernel density estimate distribution plot of abundance of protein identifications at each molecular weight in Bowman’s space aspirate (GF) and plasma. Mean of 4 experiments from each group. The black vertical line at 66 kDa indicates the molecular weight of albumin, the most prevalent plasma protein. While this is the most prevalent molecular weight region in plasma, and many proteins with greater molecular weight were identified in plasma, in Bowman’s space aspirate very few proteins were identified above this molecular weight. (D) Kernel density estimate plot of abundance of protein identifications at each theoretical isoelectric point in Bowman’s space aspirate and plasma. Mean of 4 experiments from each group. Proteins identified in Bowman’s space demonstrate a greater mean theoretical isoelectric point and are thus predicted to be more positively charged at any given pH. (E) Venn diagram of protein identifications in Bowman’s space and plasma. 200 of the 653 identifications in Bowman’s space aspirates were also identified in plasma. Representative gene symbols of plasma/Bowman’s space mutually identified proteins are shown. (F) Mass spectrometry of Bowman’s space aspirate and of lysate from surrounding cell types (proximal tubular epithelial cells [PTEC], glomerular endothelial cells [gENC], and podocytes) indicates minimal contamination of Bowman’s space aspirate with renal cell proteins. <10% of total identifications in Bowman’s space aspirate were specific to surrounding cell types.