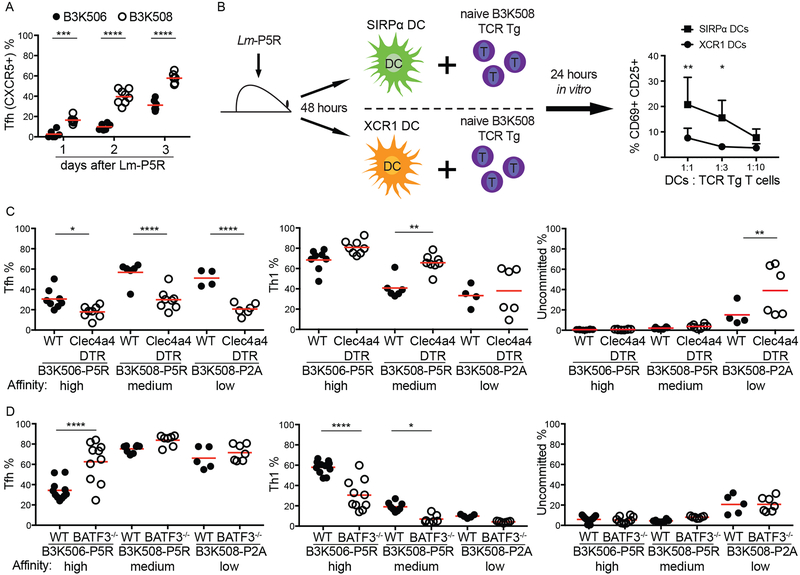
Figure 2. Effect of genetic ablation of XCR1+ or SIRP⍺+ DCs on Th differentiation.
(A) Tfh differentiation by B3K506 (filled circle, n=6) and B3K508 (empty circle, d 1 d 2 n=6, d 3 n=8) cells in the initial three d following Lm-P5R infection. (B) Experimental model and resulting mean frequencies ± SD of CD69+ CD25+ B3K508 cells after one d of co-culture with SIRP⍺+ (square, n=3) or XCR1+ (circle, n=3) DCs from Lm-P5R infected mice two d post-infection. (C) Frequency of Tfh, Th1, or uncommitted cells among B3K506 or B3K508 TCR Tg cells in DT-treated WT (filled circle) or Clec4a4DTR (empty circle) mice three d after Lm-P5R (B3K506 WT n=8, Clec4a4DTR n=8; B3K508 WT n=6, Clec4a4DTR n=8) or Lm-P2A (B3K508 WT n=4, Clec4a4DTR n=6) infection. (D) Frequency of Tfh, Th1, or uncommitted cells among B3K506 or B3K508 cells in WT (filled circle) or Batf3−/− (empty circle) mice three d after Lm-P5R (B3K506 WT n=11, Batf3−/− n=7; B3K508 WT n=8, Batf3−/− n=7) or Lm-P2A (B3K508 WT n=5, Batf3−/− n=7) infection. The bars in A, C, and D represent the mean. Pooled data from two or three independent experiments are shown. Spleens were analyzed for the depicted experiments. Two-way ANOVA was used to determine significance for B and one-way ANOVA with Sidak’s multiple comparison test was used to determine significance for A, C and D. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.