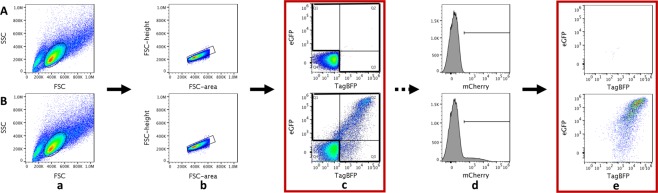
Figure 2.
Assay gating strategies. The top panels (A) show untransfected cells. The bottom panels (B) show cells transfected with the vector constructs shown in Fig. 1b. 293T/17 cells for analysis were identified by gating first on a forward-scatter versus side-scatter plot (a) followed by an additional gate for single cells constructed on a forward-scatter area versus forward-scatter height plot (b). The fluoresence intensity (eGFP versus TagBFP) of each individual cell is displayed, representing the two markers carried on the HIV-derived construct (c). Untransfected cells were used to define the double-negative population which was excluded from further analysis. Cells which expressed the HIV-derived construct (i.e. Q1, Q2 and Q3 - eGFP-positive or TagBFP-positive or both) were used for analysis. In other experiments utilizing the MSCV-derived construct carrying a third fluorescent marker, an alternative analysis could be used. In addition to gating on single cells expressing the HIV-derived construct, a gate was created including cells expressing the MSCV-derived construct as well (i.e. mCherry positive) (d). Finally, the population of cells expressing both the HIV-derived construct (i.e. Q1, Q2, or Q3 from c) and the MSCV-derived construct (i.e. mCherry positive from d) was used for analysis (e). Red boxes denote gates used for calculation of functional activity.