Abstract
Long non-coding RNAs (lncRNAs) have recently emerged as new regulatory molecules with diverse functions in regulating gene expression and significant roles in the immune response. However, the function of many unknown lncRNAs is still unclear. By studying the regulatory effect of daidzein (DA) on immunity, we identified a novel lncRNA with an immune regulatory function: lncRNA- XLOC_098131. In vivo, DA treatment upregulated the expression of lncRNA- XLOC_098131, FOS, and JUN in chickens and affected the expression of activator protein 1 (AP-1) to regulate MAPK signaling, Toll-like receptor signaling, and related mRNA expression. It also enhanced macrophage activity and increased the numbers of blood neutrophils and mononuclear cells, which can improve the body's ability to respond to stress and bacterial and viral infections. Furthermore, DA treatment also reduced B lymphocyte apoptosis and promoted the differentiation of B lymphocytes into plasma cells, which in turn resulted in the production of more immunoglobulins and the promotion of antigen presentation. In vitro, using HEK293FT cells, we demonstrated that mir-548s could bind to and decrease the expression of both FOS and lncRNA- XLOC_098131. LncRNA- XLOC_098131 served as a competitive endogenous RNA to stabilize FOS by competitively binding to miR-548s and thereby reducing its inhibitory effect of FOS expression. Therefore, we concluded that the novel lncRNA XLOC_098131 acts as a key regulatory molecule that can regulate the Toll-like receptor signaling pathway and related immune function by serving as a competitive endogenous RNA to stabilize FOS mRNA expression.
Keywords: competitive endogenous RNA, daidzein, immunoglobulins, lymphocytes, miR-548s, MAPK signaling, novel lncRNAs, Toll-like receptor pathway
Introduction
As a template RNA molecule, mRNA plays an important role in DNA genetic information transmission and protein synthesis (1). However, with advancements in research technologies, researchers have identified that mRNA accounts for only ~1.2% of transcripts, whereas non-coding RNA (ncRNA) in the transcriptome accounts for as much as 98%, which suggests that ncRNA might perform rich biological functions in organisms (2). Long non-coding RNAs (lncRNAs), which are non-coding RNA transcripts that are longer than 200 nucleotides and lack apparent open reading frames, play important roles in many cellular biological processes, including cell cycle progression, apoptosis, development, muscle differentiation, and immune regulation (3), and participate mainly in epigenetic, transcriptional, and posttranscriptional regulation (4). LncRNAs can regulate epigenetic processing by affecting chromatin reprogramming, DNA methylation and histone modification (5). In addition, lncRNAs mediate gene silencing by recruiting chromatin-modifying complexes toward target genes through sequence-specific binding (6). LncRNAs, such as Kcnq1ot1 and Air, which map to the Kcnq1 and Igf2r imprinted gene clusters, respectively, mediate the transcriptional silencing of multiple genes by interacting with chromatin and recruiting the chromatin-modifying machinery (7). Furthermore, lncRNAs can regulate gene expression at the transcriptional and posttranscriptional levels. The depletion of several ncRNAs leads to decreased expression of their neighboring protein-coding genes, including the master regulator of hematopoiesis, SCL (also called TAL1), Snai1 and Snai2 (8). After transcription, lncRNAs can regulate gene expression as competitive endogenous RNAs (ceRNAs) (5, 9, 10).
LncRNAs can participate in immune cell differentiation and the regulation of related processes. In CD8(+) T cells, some lncRNAs overlap with miRNAs and siRNAs, which suggests that lncRNAs act by being processed into smaller molecules that play a key role in the acquired immune response (11). The relationship between lncRNAs and dendritic cell differentiation was recently explained as follows: lnc-DC directly binds STAT3 in the cytoplasm and thus promotes STAT3 phosphorylation on tyrosine-705 by preventing STAT3 from binding to and being dephosphorylated by SHP1, and this phosphorylation activates the tyrosine kinase JAK/STAT signaling pathway and regulates dendritic cell differentiation (12). Recent studies revealed that in the immune system, in addition to directly regulating immune cells, lncRNAs can control inflammation by directly acting on inflammatory factors, and other studies found that lncRNAs can participate in the epigenetic modification of inflammation-related genes (13). Stimulation with lipopolysaccharide significantly increases the expression of lnc-IL7R in cells, and the expression of this lncRNA is associated with epigenetic regulation (14). In addition, lncRNA-Cox2 plays a key role in the regulation of IL-6 expression induced by Toll-like receptor stimulation (15). Multiple studies have shown that lncRNAs can be used as ceRNAs to indirectly regulate immunity-related mRNA expression through competitive binding to miRNA (16, 17). For example, let-7e regulates the inflammatory response of vascular endothelial cells by ceRNA crosstalk (18). The lncRNA XLOC_008466 regulates oncogenes in human lung cancer cells by targeting miR-874 (19), and lncRNAs play a positive role as competitive endogenous RNAs in gastric cancer (20). The involvement of lncRNAs in immune regulation is complicated, and many key immune regulatory lncRNAs have not yet been identified. Therefore, our study aimed to reveal some key novel lncRNAs in immune regulation.
Daidzein (4′,7-dihydroxyisoflavone) is a naturally occurring isoflavonic phytoestrogen belonging to the non-steroidal estrogen family and is mainly derived from leguminous plants (21). DA is also a major bioactive ingredient in the traditional Chinese medicine Gegen, which is frequently used for the treatment of fever, acute dysentery, diabetes, cardiac dysfunctions, and liver injury, among other conditions (22). The chemical structure of DA is similar to that of 17β-estradiol (E2), and DA can selectively bind different estrogen receptors (ER) at different affinities to regulate the recruitment of co-repressors and co-activators and affect ER signaling (23). Therefore, DA exerts protective effects against some diseases that are linked to the regulation of estrogen, such as breast cancer, osteoporosis, diabetes, and cardiovascular diseases (24). DA also has several other biological activities that are independent of the ER, such as anti-inflammation and anticancer activities and protection of the skin and nerves. These beneficial effects are mainly due to the regulation of the immune response (25). DA has been shown to precisely orchestrate processes related to the regulation of the secretion of immune molecules, the proliferation and differentiation of immune cells, and immune signal pathways. Feeding mice DA after the onset of experimental allergic encephalomyelitis reduced the secretion of interferon-3 and interleukin-12, enhanced interleukin-10 production, suppressed lymphocyte proliferation, and decreased cytotoxicity (26). Studies have shown that under conditions of LPS-induced inflammation, DA could inhibit inflammatory hyperreactivity and eliminate inflammation by inhibiting the differentiation of B lymphocytes (27). In addition, mice treated with DA exhibited increased percentages of CD4(+) and CD28(+) T cells, and DA treatment regulated B lymphopoiesis and decreased the mRNA levels of RANKL in B220(+) cells (28). DA can affect the immune response by regulating the Toll-like and NF-κB pathways. We thus attempted to identify key lncRNAs involved in immune regulation by studying the daidzein-mediated changes in lncRNA expression associated with the regulation of the immune response (29, 30).
Numerous studies have shown that plant extracts not only regulate the expression of coding genes but also exert regulatory effects on non-coding genes (31–34). Although these studies highlight the relevance of lncRNAs in the immune regulation process, the precise molecular mechanisms remain largely unelucidated. Furthermore, the role of lncRNAs involved in the regulation of the immune response to DA remains unknown. In this study, we treated egg-laying hens with DA, screened for key mRNAs and lncRNAs related to immune regulation using transcriptome technology, and performed cell experiments to explore the specific molecular biological mechanisms underlying the regulation of immune responses by lncRNAs.
Materials and Methods
Materials
The DA used in this study was synthetically produced by the Kai Meng Co. (Xi An, Shanxi, China) Chemical Plant with a purity of 99.9%.
Feeding Experimental Design and Bird Management
The experimental animal procedures were approved by the China Agricultural University Animal Care and Use Committee (Beijing, China, permit number SYXK20130013). The experiment was performed with laying broiler breeder hens housed at a commercial farm (Zhuozhou, China) under standard conditions. After a 2-week acclimation period, a total of 480 57-week-old Ross 308 laying broiler breeder hens were allocated to two treatment groups: the DA-deficient group (DAD), which the diet is specially formulated lacking DA, and the DA-supplemented group (DS). Each treatment was replicated eight times, and each replicate included 30 broiler breeder hens. The hens were fed a nutritionally balanced corn-miscellaneous meal (CSCM) with DA added at 0 and 20 mg/kg for 8 weeks. The CSCM diets were formulated to meet the nutrient requirements of laying broiler breeders according to the NRC guidelines (1994) (Table 1).
Table 1.
Composition and nutritional levels in the experimental diets of hens.
| Ingredient (%) | DAD group diet | DS group diet |
|---|---|---|
| Corn | 68.99 | 68.99 |
| Soybean meal | 4 | 4 |
| Corn protein | 9.15 | 9.15 |
| De-gossypol cottonseed protein | 6 | 6 |
| Limestone | 7.76 | 7.76 |
| Soybean oil | 0.5 | 0.5 |
| Dicalcium phosphate | 2.09 | 2.09 |
| NaCl | 0.35 | 0.35 |
| aTrace mineral premix | 0.3 | 0.3 |
| Choline chloride (50%) | 0.12 | 0.12 |
| Mycotoxin adsorbent | 0.1 | 0.1 |
| DL-methionine | 0.0515 | 0.0515 |
| bVitamin premix | 0.035 | 0.035 |
| Santoquin | 0.03 | 0.03 |
| Phytase | 0.016 | 0.016 |
| 4% Flavomycin | 0.015 | 0.015 |
| Lysine•HCl (8%) | 0.373 | 0.373 |
| Threonine | 0.0664 | 0.0664 |
| Tryptophan | 0.0481 | 0.0481 |
| Daidzein | 0 | 20 ppm |
| Total | 100 | 100 |
| Avian metabolic energy (MC/kg) | 2.83 | 2.83 |
| Crude protein (%) | 16.1 | 16.1 |
| Calcium (%) | 3.48 | 3.48 |
| Total phosphorus (%) | 0.678 | 0.678 |
| Available phosphorus (%) | 0.47 | 0.47 |
| Methionine (%) | 0.34 | 0.34 |
| Lysine (%) | 0.805 | 0.805 |
| Met+Cys (%) | 0.626 | 0.626 |
| Threonine (%) | 0.6 | 0.6 |
| Tryptophan (%) | 0.18 | 0.18 |
Supplied the following per kg complete diet: Cu, 8 mg; Zn, 75 mg; Fe, 80 mg; Mn, 100 mg; Se, 0.15 mg; I, 0.35 mg.
Supplied the following per kg complete diet: vitamin A, 12,500 IU; vitamin D3, 2,500 IU; vitamin E, 30 IU; vitamin K3, 2.65 mg; thiamine, 2 mg; riboflavin, 6 mg; vitamin B12, 0.025 mg; biotin, 0.0325 mg; folic acid, 1.25 mg; pantothenic acid, 12 mg; niacin, 50 mg.
Sample Collection and Chemical Analysis
At the 4 and 8th weeks of the experiment, one chicken per replicate was selected and deprived of food for 8 h. One blood sample was collected from the wing vein of each replicate chick into vacuum blood collection tubes, and the serum was centrifuged at 3000 × g for 15 min and stored at −20°C until use for the detection of immunoglobulins (Igs). Another blood sample was collected from the wing vein of each replicate chick into vacuum blood collection tubes (with heparin sodium) for assessing lymphocyte proliferation and percentages. At the 8th week of the experiment, one chicken from each replicate was slaughtered. One liver sample was immediately collected from each replicate for measuring gene expression, and the samples were frozen in liquid nitrogen and stored in a freezer at −80°C.
Serum Immunoglobulin Levels
The serum IgM and IgA levels were determined using a commercial ELISA kit (IDEXX laboratories Inc., Westbrook, Maine, USA) according to the manufacturer's recommended protocol.
Lymphocyte Classification and Proliferation
Peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density centrifugation. Briefly, heparinized blood was diluted with Hank's balanced salt solution at a ratio of 1:1 (no calcium and no magnesium, Life Technologies, Burlington, Vermont, USA) and carefully layered on top of Histopaque 1077 (Sigma-Aldrich Corporation, Burlington, Vermont, USA) in a 10-mL centrifuge tube at a 2:1 ratio. After centrifugation for 30 min at 3,000 rpm and 20°C, the PBMCs at the plasma-Ficoll interface were collected and washed three times with cold RPMI-1640 medium (containing 5.0% inactivated fetal bovine serum, 0.0599 mg/mL penicillin, 100 μg/mL streptomycin, and 24 mM HEPES) by centrifugation at 1,800 rpm and 4°C for 10 min. The cell counts and viability were evaluated by trypan blue staining. The lymphocytes were then mixed with CD3 (SPRD), CD4 (FITC), and CD8 (RPE) antibodies or Bu-1 (RPE) antibodies, and the cells were then incubated in a water bath at 37°C for 30 min, washed twice with Hanks solution and fixed with 3% paraformaldehyde. The results are expressed as percentages. The proliferative responses of T and B cells after stimulation with concanavalin A (ConA, 45 μg/mL) and LPS (25 μg/mL), respectively, were determined by an MTT assay. ConA from Canavalia ensiformis (C2010) and LPS from Escherichia coli (L2880) were both obtained from Sigma-Aldrich Corporation. The results are expressed as SI values (35).
Blood Routine Examination
The routine blood examination was performed at Xiyuan Hospital, Beijing, China.
Next-Generation Sequencing (NGS)
Total RNA samples for sequencing were purified from 20 mg of tissue samples from eight chickens (four replicates in each treatment group) using the RNeasy Fibrous Tissue Mini messenger RNA (mRNA) extraction kit (Qiagen, Hilden, Germany) following the manufacturer's recommendations. The concentration and purity of total RNA were determined using a UV/Vis spectrophotometer (ACTGene, New Jersey, USA) at 260 nm, and the sample integrity was evaluated through a microfluidic assay using a Bioanalyzer system (Agilent Technologies, Inc., Santa Clara, California, USA). Only high-quality RNA extracts [RNA integrity number (RIN) ≥ 8] were used to pool equal amounts of RNA per chicken within each treatment group, and NGS data were obtained from the pooled RNA samples from each group to ensure the most robust transcriptome. Complementary DNA (cDNA) libraries for RNA sequencing (RNA-Seq) were constructed using a TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, California, USA), and RNA-Seq analysis was performed to identify transcriptional changes using a MiSeq instrument (Illumina) with paired-end libraries (CapitalBio, http://cn.capitalbio.com/) (36). Four replicates from each treatment were analyzed independently for library synthesis and sequencing, and the quality of the raw reads was assessed using FastQC (Version 0.10.1). Adapters, low-quality reads at the 3′ end, reads with fuzzy N bases, ribosomal RNA (rRNA), sequences shorter than 20 nt and low-quality reads (those with a Q < 20) were trimmed with the FASTX clipper (Version 0.0.13). All the double-end reads in eight samples from two treatment groups were separately aligned to the chicken reference genome (Gallus_gallus-5.0, version 81, Ensembl) using the spliced mapping algorithm in TopHat2 (version: 2.0.9) (37). Unless stated otherwise, all programs were run with the default parameters. The number of reads equivalent to mapped reads [reads per kilobase per million mapped reads (RPKM)] was used to normalize the expression of each gene. The quality of the obtained data was assessed based on the presence and abundance of contaminating sequences, the average read length, and the GC content. With the exception of the microarray design, the NGS experiment conformed to the MIAME guidelines (38).
Bioinformatics Analysis
All data processing steps for differential gene expression evaluation were performed using the Cuffdiff software package (DNASTAR, Madison, Wisconsin, USA). Further analysis was conducted only using genes that demonstrated a >1.5-fold change or a <0.7-fold change in expression between the groups, as demonstrated by t-tests with an adjusted P-value ≤ 0.05. A Benjamini and Hochberg test (with an error rate of 0.05) and a false discovery rate correction test (FDR < 0.05) were used to adjust the P-values. The biological mechanisms underlying the DEGs were investigated using DAVID v. 6.7 software (the Database for Annotation, Visualization, and Integrated Discovery) (39) (http://david.abcc.ncifcrf.gov/). The sets of genes were uploaded using official gene IDs. Finally, the molecular interaction networks of the DEGs were investigated using OmicsBean (http://www.omicsbean.cn) and Cytoscape v. 3.1.0 software (http://www.cytoscape.org/) (40) as a complementary and more comprehensive approach for identifying the central hub genes. The results were visualized using the ClueGO v. 2.1.1. application in Cytoscape, which creates clusters of functionally related genes using the following GO databases: KEGG, Wiki Pathways, REACTOME, and GO.
LncRNA Identification
To identify novel reliable lncRNA models, we considered only multiexonic transcripts and filtered them through the following highly stringent criteria: (1) size selection: only transcripts with ≥200 bp were kept; (2) read coverage threshold: transcripts with ≤ 3 reads were removed from our dataset; (3) open reading frame (ORF) filter: transcripts with a predicted ORF longer than 100 aa were removed; (4) known protein domain filter: transcripts were aligned to the Pfam and Swiss-Protein databases to eliminate transcripts with significant homology to known protein domains (41); and (5) protein-coding-score test: both the Coding-Non-Coding Index (CNCI) and the CPC were used to evaluate the coding potential of the candidate lncRNAs (42, 43). Further analysis was conducted only with the lncRNAs that showed >1-log2 (fold change) or <-1-log2 (fold change) differential expression between the dietary groups, as demonstrated through t-tests with an adjusted P-value ≤ 0.05.
RNA Fluorescence in situ Hybridization (RNA FISH)
Cy3-labeled lncRNA- XLOC_098131 probes were obtained from RiboBio (Guangzhou, China). RNA FISH was performed using a fluorescent in situ hybridization kit (RiboBio) following the manufacturer's instructions.
5′- and 3′-Rapid Amplification of cDNA Ends (RACE)
To determine the full-length sequence of lncRNA- XLOC_098131, RACE experiments were performed using the SMARTer RACE cDNA Amplification Kit (Clontech, Palo Alto, California, USA) following the manufacturer's recommended protocol. The gene-specific primers used for 5′ and 3′ RACE were GTTCTGCAGGAGAGCAGCAG and AAGTGGCCTGAGCTGGAGTC, respectively.
Constructs
The full length of lncRNA- XLOC_098131 contains a 3′UTR luciferase reporter WT plasmid named pHS-AVC-LW406 (psi-SV40 promoter-hRluc- lncRNA- XLOC_098131-HSV TK promoter-hluc) (Figures S1A,B); a mutant expression plasmid of lncRNA- XLOC_098131 without the predicted miR-548s-binding sites named pHS-AVC-LW407 [psi-SV40 promoter-hRluc- lncRNA- XLOC_098131 (mutation)-HSV TK promoter-hluc] (Figures S1C,D), mir-548s overexpression plasmids named pHS-AMR-LW010 (pZDonor_hef1a-EYFP-T2A-puro-hsa-mir-548ah) (Figure S1E), pHS-AMR-LW011 (pZDonor_hef1a-EYFP-T2A-puro-hsa-mir-548ay) (Figure S1F), and pHS-AMR-LW012 (pZDonor_hef1a-EYFP-T2A-puro-hsa-mir-548e) (Figure S1G), and a control mi plasmid named pHS-AMR-LW013 (pZDonor_hef1a-EYFP-T2A-puro-control miRNA) (Figure S1H). These plasmids were constructed by Hesheng Gene Company (Beijing, China) to test the ability of lncRNA- XLOC_098131 to bind to mir-548s. The full length of FOS contains a 3′UTR luciferase reporter WT plasmid named pHS-AVC-LW480 [psi-SV40 promoter-hRluc-Fos gene part (hsa-miRNA-548ay-3p target)-HSV TK promoter-hluc] (Figure S2A) and a mutant expression plasmid of FOS without the predicted miR-548s-binding sites named pHS-AVC-LW481 [psi-SV40 promoter-hRluc-Fos gene part mutation (hsa-miRNA-548ay-3p target mutation)-HSV TK promoter-hluc] (Figure S2B), and these were constructed by Hesheng Gene Company (Beijing, China) to test the binding between FOS and mir-548s. An FOS luciferase vector named pHS-AVC-LW482 (psi-SV40 promoter-hRluc-Fos gene part (hsa-miRNA-548ay-3p target)-HSV TK promoter-hluc) (Figure S2C), an lncRNA- XLOC_098131 overexpression plasmid and a mutant overexpression plasmid of lncRNA- XLOC_098131 without predicted miR-548s-binding sites (Figure S2D) were constructed by Hesheng GenePharma Company (Beijing, China) to verify that lncRNA- XLOC_098131 regulates FOS expression by competitively binding to miR-548s. We also constructed a mutant expression plasmid of lncRNA- XLOC_098131 without predicted miR-1180s-binding sites named pHS-AVC-LW251 [psi-SV40 promoter-hRluc- lncRNA- XLOC_098131 (mutant)-HSV TK promoter-hluc] (Figure S3B) and mir-1180s overexpression plasmids named pHS-AMR-ZQ011 (pZDonor_hef1a-EYFP-T2A-puro-hsa-mir-1180-5P) and pHS-AMR-ZQ012 (pZDonor_hef1a-EYFP-T2A-puro-hsa-mir-1180-3P) (Figure S3A) to test the binding between lncRNA- XLOC_098131 and mir-1180s.
Luciferase Assays
For the luciferase reporter assays, HEK293FT cells were seeded into 96-well plates in triplicate and transfected with the WT lncRNA- XLOC_098131 construct pHS-AVC-LW406, the mutant lncRNA- XLOC_098131 construct pHS-AVC-LW407, psiCHECK-2 with the mir-548s overexpression plasmids pHS-AMR-LW010, pHS-AMR-LW011, and pHS-AMR-LW012, or the control miR plasmid pHS-AMR-LW013. Forty-eight hours after transfection, the luciferase activities were measured using a Dual-Luciferase® Reporter Assay System (Promega). HEK293FT cells were then seeded into 96-well plates in triplicate and transfected with the WT FOS construct pHS-AVC-LW480, the mutant FOS construct pHS-AVC-LW481, psiCHECK-2 with the mir-548s overexpression plasmids pHS-AMR-LW010, pHS-AMR-LW011, and pHS-AMR-LW012, or the control mi plasmid pHS-AMR-LW013. Forty-eight hours after transfection, the luciferase activities were measured. Additionally, HEK293FT cells were seeded into 96-well plates in triplicate and transfected with the FOS luciferase vector pHS-AVC-LW480 or control psiCHECK with pcDNA3.1, lncRNA- XLOC_098131, lncRNA- XLOC_098131-mut or lncRNA- XLOC_098131+mir-548s. Forty-eight hours after transfection, the luciferase activities were measured. In addition, the luciferase assays of JUN and lncRNA- XLOC_098131 were the same as those used in the FOS assay.
Quantitative Real-Time Reverse Transcription PCR
The expression levels of various genes were analyzed by qRT-PCR. The gene-specific primer sequences are shown in Table 2, and the analysis was performed on a 7500-fluorescence detection system (Applied Biosystems, Foster City, California, USA) using a commercial SYBR-Green PCR kit (Takara Bio Inc. Foster City, California, USA). According to the manufacturer's recommended protocol, the following PCR conditions were employed: 95°C for 30 s and 40 cycles of 95°C for 5 s and 60°C for 34 s. In addition, melting curve analyses and subsequent agarose gel electrophoresis of the PCR products were conducted to confirm the amplification specificity. The relative gene expression data were analyzed using the 2−ΔΔCt method.
Table 2.
qRT-PCR primers for gene analysis.
| Target | Primer sequence (5′-3′)a | Product size, bp |
|---|---|---|
| FOS | F: CGGGAGAGGAACAAGATGG R: TCCTTCAGCAGGTTGGCTAT |
137 |
| CTSK | F: AGTCTGCCCTCCTTCCAGTT R: TGCTTGGTGCCCTTCTGT |
119 |
| BDKRB1 | F: GTGTATCGACGCCATCTGTG R: GACAGCCAGGTTCACAAGGT |
118 |
| KL | F: TCCAGGAACGACCAAGAAGT R: CAACGCTGTTTCTCTGGTGA |
130 |
| CYR61 | F: CTGCAGAGCACAGTCTGAGG R: CAGCCCACAGCTCCATCTAT |
120 |
| CDH2 | F: GCCCATTGACTTTGAGACCA R: GACACGGTTGCTGTTGACTG |
111 |
F refers to forward, and R refers to reverse.
Statistical Analysis
When a significant difference was observed between treatments, the individual treatment means were compared with Duncan's multiple comparison using SPSS Version 18.0. A P-value < 0.05 was considered to indicate statistical significance, and P < 0.1 was considered to indicate a trend toward statistical significance.
Results
Serum Immunoglobulin Levels, Peripheral Blood Lymphocyte Classification, and Lymphocyte Proliferation in Hens
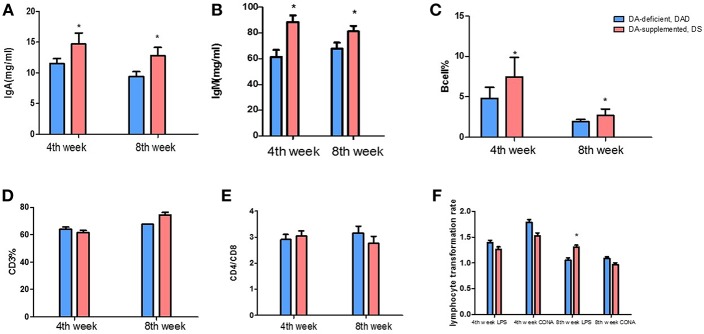
Compared with the DA-deficient (DAD) group, DA supplementation at a dose of 20 mg/kg in hens increased (P < 0.05) the serum IgA and IgM levels on the 4 and 8th weeks of the experiment, as shown in Figures 1A,B. The effects of DA treatment on peripheral blood lymphocytes are also shown in Figures 1C–F. DA supplementation at a dose of 20 mg/kg increased (P < 0.05) the ratio of B cells on the 4 and 8th weeks of the experiment (Figure 1C), but no significant effect (P > 0.05) on the percentage of CD3 (Figure 1D) and the CD4/CD8 ratio was observed (Figure 1E). We further examined the lipopolysaccharide (LPS)- and concanavalin A (CON A)-mediated stimulation of the lymphocyte proliferation rate and found that the DA treatment increased (P < 0.05) the proliferative responses of B cells on the 8th week of the experiment, as shown by changes in the LPS stimulation index (SI) values obtained in vitro using peripheral blood (Figure 1F). In contrast, no difference in lymphocyte proliferation was observed following CON A stimulation.
Figure 1.
Effects of dietary daidzein supplementation of hens on the serum immunoglobulin levels and peripheral blood lymphocyte classification and lymphocyte proliferation. (A) Serum immunoglobulin A levels of hens at the 4 and 8th weeks of the experiment. (B) Serum immunoglobulin M levels of hens at the 4 and 8th weeks of the experiment. (C) Peripheral blood B lymphocyte percentages of hens at the 4 and 8th weeks of the experiment. (D) Peripheral blood CD3(+) T cell percentages of hens at the 4 and 8th weeks of the experiment. (E) Peripheral blood CD4/CD8 T cells of hens at the 4 and 8th weeks of the experiment. (F) In vitro lymphocyte proliferation in hens at the 4 and 8th weeks of the experiment. Data information: We induced T and B lymphocyte cell multiplication in peripheral blood lymphocytes through concanavalin A and lipopolysaccharide stimulation, respectively. LPS, lipopolysaccharide stimulus index; CON A, concanavalin A stimulus index. The results are expressed as the means ± SEMs. Bars labeled with “*” are significantly different (P < 0.05).
Routine Blood Examination
The routine blood examination results (Table 3) showed that on the 4th week of the experiment, the platelet count (PLT) and pressure (PCT) in the DA-supplemented (DS) group were decreased (P < 0.05) compared with those found in the DAD group, and the DS group also exhibited a slight increase (P < 0.1) in the neutrophil ratio (NEUT%). Furthermore, DA significantly increased the percentage of monocytes (MONO%) and the proportion of large unstained cells (LUC%). On the 8th week of the experiment, the MONO and LUC% in the DS group were higher (P < 0.05) than those in the DAD group, and the NEUT% showed an increasing tendency (P < 0.1). In addition, the DA treatment did not affect the white blood cell percentage (WBCP), average platelet volume (MPV), platelet distribution width (PDW), lymphocyte ratio (lymph%), eosinophilia ratio (EOS%), or basophil ratio (BASO%).
Table 3.
Effects of dietary daidzein supplementation of hens on routine blood examination results*.
| WBCP | PLT | MPV | PDW | PCT | NEUT(%) | Lymph(%) | MONO(%) | EOS(%) | LUC(%) | BASO(%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Week 4 | |||||||||||
| DAD | 76.29 | 55.38a | 41.85 | 23.65 | 0.23a | 14.23 | 79.58 | 4.34b | 0.19 | 0.66b | 0.63 |
| DS | 76.10 | 36.38b | 41.71 | 24.83 | 0.15b | 19.99 | 80.68 | 6.71a | 0.23 | 1.11a | 0.46 |
| SEM | 3.634 | 3.617 | 0.298 | 0.785 | 0.016 | 1.618 | 1.502 | 0.554 | 0.027 | 0.102 | 0.060 |
| P-value | 0.980 | 0.004 | 0.827 | 0.474 | 0.006 | 0.073 | 0.728 | 0.026 | 0.499 | 0.022 | 0.183 |
| Week 8 | |||||||||||
| DAD | 79.74 | 53.13 | 41.88 | 21.54 | 0.25 | 15.37 | 73.14 | 4.49b | 0.26 | 1.09b | 0.76 |
| DS | 83.47 | 69.38 | 36.41 | 23.05 | 0.24 | 20.65 | 70.33 | 7.29a | 0.31 | 1.73a | 0.63 |
| SEM | 4.585 | 6.772 | 1.794 | 0.689 | 0.037 | 1.577 | 2.182 | 0.555 | 0.033 | 0.155 | 0.088 |
| P-value | 0.699 | 0.243 | 0.132 | 0.295 | 0.847 | 0.095 | 0.538 | 0.006 | 0.464 | 0.034 | 0.455 |
WBCP, white blood cell percentage (%); PLT, platelet count (109/L); MPV, mean platelet volume (fL); PDW, platelet volume distribution width (%); PCT, platelet thrombocytocrit (%); NEUT (%), neutrophil ratio (%); lymph%, lymphocyte ratio (%); MONO%, monocyte ratio (%); EOS%, eosinophil ratio (%); LUC%, proportion of large unstained cells (%); BASO%, basophil ratio (%). The data are expressed as the means with SEMs and P-values.
Values with the same superscript within a column are not significantly different at P < 0.05, values with the different superscript (a,b) are significantly different at P < 0.05.
RNA-Seq Statistics
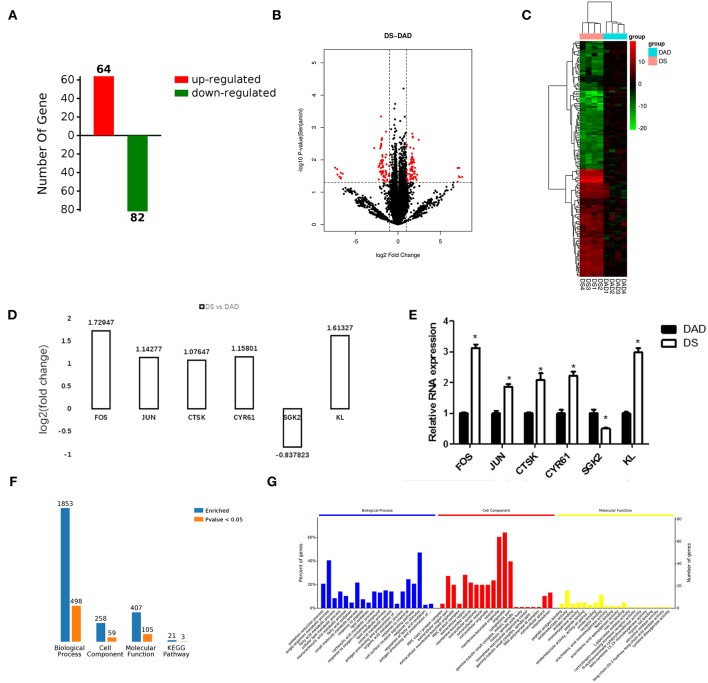
In this study, we established eight cDNA libraries from the livers of the birds in the DAD and DS groups with four replicates per group. The RNA-Seq assay generated 79,846,624–93,885,866 raw reads per library, with an average of 88,374,633 and 83,729,690 paired-end reads in the DAD and DS groups, respectively. After filtering the low-quality reads, the average numbers of clean reads in the DAD and DS groups were 82,831,728 (93.73%) and 74,604,252 (89.10%), respectively. The clean reads were used in all further analyses. Approximately 83.0% of the reads in each library were uniquely mapped to the galGal4 assembly of the chicken genome, and the average mapping rates of the DAD and DS groups were 92.7 and 93%, respectively (Table S1). As shown in Figure 2A, 146 differentially expressed genes (DEGs; 64 upregulated and 82 downregulated) were identified in the comparison of the DAD group with the DS group (FDR ≤ 0.05, fold change ≥1.5 or ≤ 0.7) using Cuffdiff software. The expression levels and fold changes of the DEGs are shown in Table S2, and the DEGs are also shown in a heat map (Figure 2B) and volcano plot (Figure 2C). Moreover, the transcriptome data of the DEGs were validated by RT-PCR (Figure 2E), and the results were consistent with the RNA-Seq results (Figure 2D).
Figure 2.
Effects of dietary daidzein supplementation of hens on differential gene expression and functional enrichment. (A) Number of differentially expressed genes (DEGs) (up/downregulated) in hens after daidzein supplementation at 20 mg/kg. (B) Heatmap of expression of DEGs in samples. DAD, daidzein-deficient group; DS, daidzein-supplemented group. DAD group samples: DAD1-4; DS group samples: DS1-4. (C) Volcano map of the expression of DEGs. (D) Fold change based on transcriptome results for six DEGs. (E) Relative RNA expression of the six genes by qRT-PCR. (F) Number of DEGs enriched in GOs and pathways. (G) Top 20 significantly enriched biological processes, cell components, and molecular functions.
Functional Categorization of DEGs and Pathway Analysis
We performed a functional enrichment analysis using the 146 DEGs, and results showed revealed 1,853, 259, 407, and 21 terms associated with biological process regulation, cell components, molecular function, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, respectively, and 498, 59, 105, and 3 significantly enriched terms in these categories, respectively (Figure 2F). The DA treatment mainly affected the GO terms: oxidation-reduction process, fatty acid metabolic process, response to peptides, response to oxygen-containing compounds, antigen processing, presentation of peptide antigens via MHC class I and other biological processes; MHC class I protein complex, extracellular region portion, extracellular membrane-bound organelle, extracellular region, membrane-bound vesicle, and other cell components; and peptide antigen binding, oxidoreductase activity, antigen binding, receptor binding, and other molecular functions (Figure 2G). A complete list of the different gene ontology (GO) terms identified in this analysis is provided in Table S3.
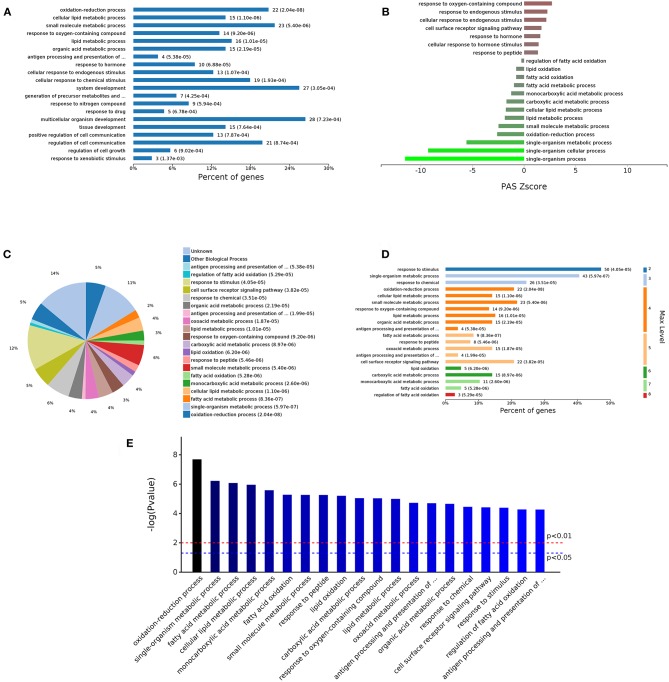
The 498 different biological process GO terms were further analyzed using OmicsBean software. According to the number of enriched genes (Figures 3A,C), the DEGs identified from the comparison of the DS group with the DAD group were mainly enriched in multicellular organism development (28 genes), system development (27 genes), and oxidation-reduction process (22 genes). According to the different levels of the GO terms (Figure 3D), most differential genes were related to stimuli (level 2 GO), and continued subdivision of the grades showed that the differential genes were involved in the organism response to multiple stimuli (level 3–8 GO). A sorting of the different GO terms based on the P-value (Figure 3E) revealed that these DEGs are most likely involved in biological processes related to antioxidation and the immune response. The PAS Z-score (Figure 3B) combines the P-value and fold change data and showed that compared with the DAD group, the DS group had significantly more immune response-related GO terms associated with enhanced immunity and significantly fewer terms related to lipid oxidation and other processes, which indicates that DS can be associated with an enhancement of the body's antioxidant capacity.
Figure 3.
Analysis of 498 different biological process GOs. (A) Percent of genes associated with enriched biological processes. (B) PAS value of enriched biological processes. (C) Expressed proteins associated with enriched biological process. (D) Levels of enriched biological processes. (E) Significantly enriched biological processes.
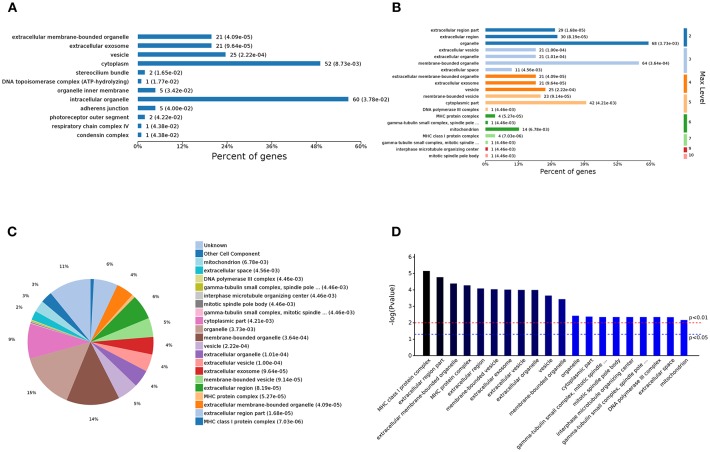
The analysis of the 59 different cell components according to the number of enriched genes (Figures 4A,C) revealed that the most abundant DEGs were enriched in intracellular organelles (60 genes) and cytoplasm (52 genes). In terms of the different GO classification levels (Figure 4B), the genes showing the highest differential expression are associated with organelles (level 2 GO), and further subdivisions showed that the DEGs are associated with membrane-bound organelles (level 3 GO). Sorting the different GO terms based on ascending P-values (Figure 4D) revealed that the genes showing the greatest differential expression are likely enriched in the cellular component of the MHC protein complex.
Figure 4.
Analysis of 59 different cell component GOs. (A) Percent of genes associated with enriched cell components. (B) Levels of enriched cell components. (C) Expressed proteins associated with enriched cell components. (D) Significantly enriched cell components.
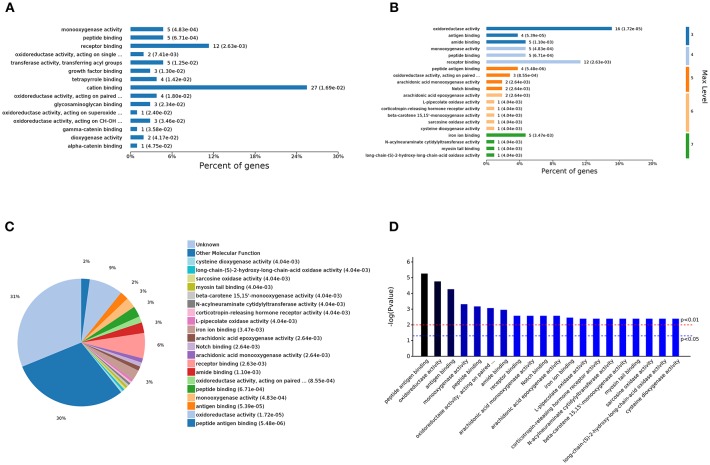
The analysis of the 105 different molecular function-related GO terms showed that according to the number of enriched genes (Figures 5A,C), the most abundantly enriched DEGs are associated with cation binding (27 genes) and receptor binding (12 genes). According to the classification of the different GO levels (Figure 5B), most DEGs are related to continued subdivision identified receptor binding (level 4 GO) and peptide antigen binding (level 5 GO). By distinguishing the difference in the P-value of the GO terms (Figure 5D), we found that the molecular functions of these DEGs are peptide antigen binding and oxidoreductase activity.
Figure 5.
Analysis of 105 different molecular function GOs. (A) Percent of genes associated with enriched molecular functions. (B) Levels of enriched molecular functions. (C) Expressed proteins associated with enriched molecular functions. (D) Significantly enriched molecular functions.
Based on the above results, it can be hypothesized that the DA treatment had an impact on the immune response, and a statistical analysis of immune regulation-related GO terms (Table 4) showed that the differential genes are involved in the positive regulation of responses to stimulus, interleukin-8 endocytosis involved viral entry into host cell, chemical stimulus, drug stimulus, endogenous stimulus, abiotic stimulus, oxidative stress, reactive oxygen species, and other immune responses. The following immunity-related GO terms were also regulated: negative regulation of B cell apoptotic processes, antigen binding, interleukin-1-mediated signaling pathway, transport of virus, regulation of MAPK cascade, positive regulation of MAP kinase activity, regulation of MAP kinase activity, Toll-like receptor 21 signaling pathway activation of MAPK activity involved in innate immune response, and receptor-mediated endocytosis of the virus by the host cell.
Table 4.
Effects of dietary daidzein supplementation of hens on DEGs used in GO clustering analysis related to the immune system.
| Gonane | GO_ID | P-value | Genes|Fold change | Count |
|---|---|---|---|---|
| Response to interleukin-8 | GO:0098758 | 6.95E-03 | EGR1|2.376 | 1 |
| Endocytosis involved in viral entry into host cell | GO:0075509 | 1.04E-02 | CAV2|0.417 | 1 |
| Cellular response to endogenous stimulus | GO:0071495 | 1.71E-05 | CAV2|0.417;ACSL1|0.614;CYR61|2.231;EGR1|2.376;JUN| 2.208;RARA|1.526;FOS|3.316;CCK1R|5.905;CHRDL1| 0.429;IRS2|1.866;KL|3.059;CRHR2|2.007;PDK4|0.421 |
13 |
| Cellular response to peptide hormone stimulus | GO:0071375 | 3.94E-04 | CAV2|0.417;IRS2|1.866;KL|3.059;CRHR2|2.007;PDK4|0.421 | 5 |
| Cellular response to chemical stimulus | GO:0070887 | 1.81E-03 | CAV2|0.417;SOD3|0.184;ACSL1|0.614;CYR61|2.231;EGR1| 2.376;JUN|2.208;PPARGC1A|0.564;RARA|1.526;FOS| 3.316;CCK1R|5.905;CHRDL1|0.429;IRS2|1.866;KL|3.059;CRHR2| 2.007;PDK4|0.421 |
15 |
| Interleukin-1-mediated signaling pathway | GO:0070498 | 4.77E-02 | EGR1|2.376 | 1 |
| Response to stimulus | GO:0050896 | 9.99E-04 | NIM1K|1.617;CAV2|0.417;CDH2|1.906;SOD3|0.184;SLC25A25| 0.627;CRCP|0.599;ACSL1|0.614;MST1R|1.705;GFRA3| 0.646;CHAC1|0.521;ENPP2|0.6;EVC|0.468;CYR61|2.231;EGR1| 2.376;JUN|2.208;PPARGC1A|0.564;TOP2A|0.53;RARA|1.526;FOS| 3.316;IGFBP1|0.446;WFDC1|0.417;GPX2|7.618;SIK1| 1.694;CCK1R|5.905;BCO1|0.596;KIAA1324|5.521;CHRDL1| 0.429;SGK2|0.559;IRS2|1.866;KL|3.059;LPAR3| 2.868;SLC6A14|0.423;CRHR2|2.007;TRPA1| 0.123;ANKRD6|0.513;EVC2|0.376;DUSP8|1.567;JAG1| 1.703;MCC|2.125;PDK4|0.421 |
40 |
| Positive regulation of response to stimulus | GO:0048584 | 3.07E-03 | CAV2|0.417;CDH2|1.906;MST1R|1.705;EVC| 0.468;CYR61|2.231;JUN|2.208;KIAA1324|5.521;KL| 3.059;LPAR3|2.868;CRHR2|2.007;ANKRD6|0.513;JAG1| 1.703 |
12 |
| Regulation of response to stimulus | GO:0048583 | 2.59E-03 | CAV2|0.417;CDH2|1.906;MST1R|1.705;CHAC1| 0.521;EVC|0.468;CYR61|2.231;EGR1| 2.376;JUN|2.208;IGFBP1|0.446;WFDC1| 0.417;KIAA1324|5.521;CHRDL1| 0.429;KL|3.059;LPAR3|2.868;CRHR2|2.007;ANKRD6| 0.513;DUSP8|1.567;JAG1|1.703;MCC|2.125 |
19 |
| Transport of virus | GO:0046794 | 3.76E-02 | CAV2|0.417 | 1 |
| Regulation of MAPK cascade | GO:0043408 | 8.82E-05 | CAV2|0.417;CDH2|1.906;MST1R|1.705;CYR61|2.231;JUN| 2.208;KL|3.059;LPAR3|2.868;ANKRD6|0.513;DUSP8|1.567 |
9 |
| Positive regulation of MAP kinase activity | GO:0043406 | 2.76E-02 | MST1R|1.705;JUN|2.208;LPAR3|2.868 | 3 |
| Regulation of MAP kinase activity | GO:0043405 | 1.10E-02 | MST1R|1.705;JUN|2.208;LPAR3|2.868;DUSP8|1.567 | 4 |
| Response to drug | GO:0042493 | 2.12E-02 | JUN|2.208;FOS|3.316;TRPA1|0.123 | 3 |
| Toll-like receptor 21 signaling pathway | GO:0035682 | 4.10E-02 | JUN|2.208 | 1 |
| Activation of MAPK activity involved in innate immune response | GO:0035419 | 4.43E-02 | JUN|2.208 | 1 |
| Cellular response to reactive oxygen species | GO:0034614 | 3.40E-02 | SOD3|0.184;FOS|3.316 | 2 |
| Cellular response to oxidative stress | GO:0034599 | 1.58E-02 | SOD3|0.184;PPARGC1A|0.564;FOS|3.316 | 3 |
| Cellular response to hormone stimulus | GO:0032870 | 2.94E-05 | CAV2|0.417;ACSL1|0.614;JUN|2.208;RARA| 1.526;CCK1R|5.905;IRS2|1.866;KL|3.059;CRHR2| 2.007;PDK4|0.421 |
9 |
| Receptor-mediated endocytosis of virus by the host cell | GO:0019065 | 1.04E-02 | CAV2|0.417 | 1 |
| Response to endogenous stimulus | GO:0009719 | 1.66E-05 | CAV2|0.417;ACSL1|0.614;CYR61|2.231;EGR1| 2.376;JUN|2.208;RARA|1.526;FOS| 3.316;CCK1R|5.905;CHRDL1|0.429;IRS2| 1.866;KL|3.059;CRHR2|2.007;JAG1| 1.703;PDK4|0.421 |
14 |
| Response to abiotic stimulus | GO:0009628 | 2.83E-02 | SOD3|0.184;EGR1|2.376;JUN|2.208;FOS|3.316;SIK1|1.694;TRPA1|0.123 | 6 |
| Response to mechanical stimulus | GO:0009612 | 6.05E-03 | JUN|2.208;FOS|3.316;TRPA1|0.123 | 3 |
| Response to external stimulus | GO:0009605 | 1.69E-03 | SLC25A25|0.627;MST1R|1.705;GFRA3|0.646;ENPP2| 0.6;JUN|2.208;PPARGC1A|0.564;FOS| 3.316;WFDC1|0.417;SIK1|1.694;KIAA1324| 5.521;CRHR2|2.007;TRPA1| 0.123;PDK4|0.421 |
13 |
| Response to oxidative stress | GO:0006979 | 2.44E-03 | SOD3|0.184;PPARGC1A|0.564;FOS|3.316;GPX2|7.618;TRPA1|0.123 | 5 |
| Antigen binding | GO:0003823 | 5.39E-05 | LOC100859408|0.197785696238; ENSGALG00000004772|0.374843424568; LOC768350|0.376374293512; ENSGALG00000027944|0.511967097185 | 4 |
| Negative regulation of B cell apoptotic process | GO:0002903 | 2.75E-02 | IRS2|1.866 | 1 |
| Response to reactive oxygen species | GO:0000302 | 8.32E-03 | SOD3|0.184;FOS|3.316;TRPA1|0.123 | 3 |
Through a KEGG pathway cluster analysis, we also found that the DEGs identified from the comparison of the DAD group with the DS group regulated the Toll-like receptor signaling pathway, salmonella infection pathway and herpes simplex infection pathway (Figure 6A). A correlation analysis (Figure 6B) showed that the DEGs are mainly involved in the Toll-like receptor signaling pathway, salmonella infection, herpes simplex infection, MAPK signaling pathway, pyruvate metabolism, cysteine and methionine metabolism, and cardiac muscle contraction, and the key genes regulating these processes are JUN, FOS, EGR1, IRS2, CTSK, and CYR61. Furthermore, an analysis of the most significant Toll-like receptor signaling pathway in KEGG (Figure 6C) revealed that the CTSK (upstream) gene in the pathway and the dimer AP-1, which is composed of proteins encoded by the JUN and FOS (downstream) genes in the pathway, play an important role in DA-mediated regulation of immunity. AP-1 can respond to a variety of stimuli, including cytokines, growth factors, stress, and bacterial and viral infections, by regulating the expression of downstream genes. This finding is consistent with the results of the GO analysis, which showed that DA addition mainly enhanced the body's ability to respond to multiple stimuli. Therefore, FOS and JUN are key genes that enable the DA-mediated enhancement of the immune response and innate immunity in hens.
Figure 6.
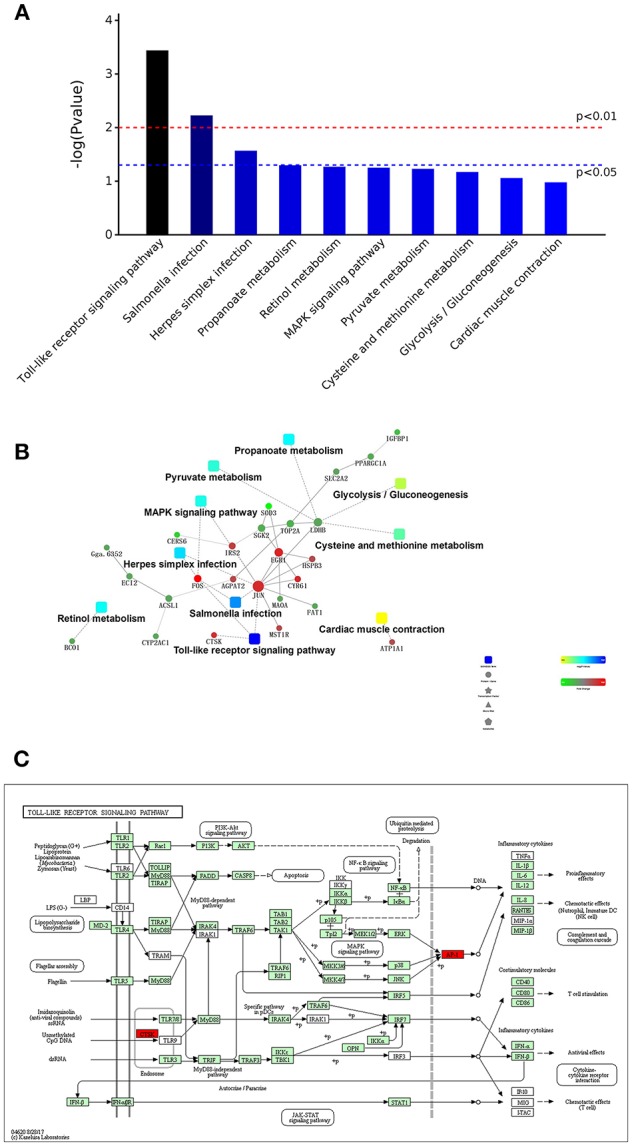
KEGG pathway cluster analysis and correlation analysis of DEGs. (A) Distribution of enriched KEGG pathways. (B) Correlation analysis of DEGs. (C) Daidzein treatment regulated the Toll-like signaling pathway. The key factors are marked in red.
Differential lncRNA Selection
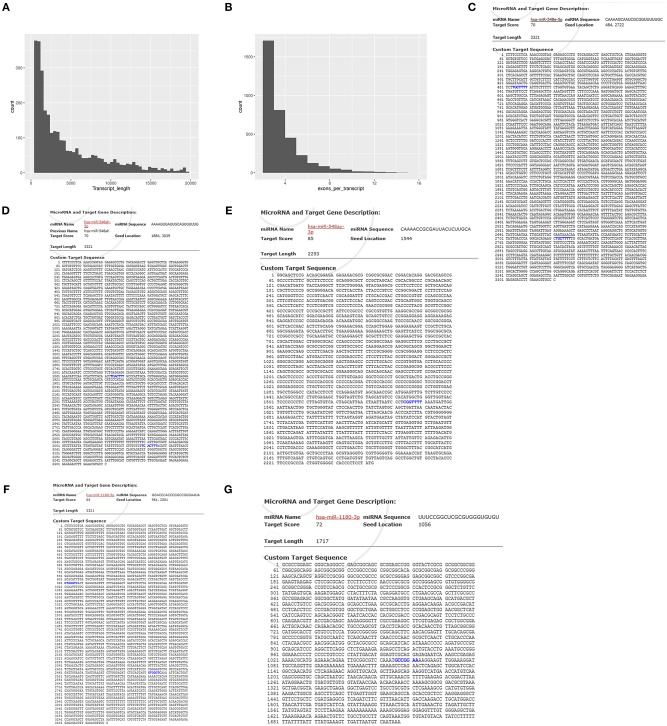
We performed a lncRNA screen of the DS and DAD groups and identified 1,095 lncRNAs (Table S4). An analysis of the transcript lengths and transcript exons of all lncRNAs (Figures 7A,B) revealed that the frequency of these lncRNA transcripts was mainly concentrated in 200–5,000 units and that the number of transcript exons mainly ranged from 2 to 4. LncRNAs that demonstrated differential expression >1-log2 (fold change) or <-1-log2 (fold change) between the two groups, as determined through t-tests with an adjusted P-value ≤ 0.05, were selected. In the DS group, nine and 22 lncRNAs were upregulated and downregulated, respectively compared with their levels in the DAD group (Table 5).
Figure 7.
Transcript length and transcript exons of lncRNAs and predicted binding sites for gene sequences and miRNAs. (A) Transcript length of lncRNAs. (B) Exon numbers of lncRNA transcripts. (C) Predicted binding sites of lncRNA- XLOC_098131 and miR-548s (miR-548e-5p). (D) Predicted binding sites of lncRNA- XLOC_098131 and miR-548s (miR-548 ah−5p). (E) Predicted binding sites of FOS and miR-548s (miR-548ay-3p). (F) Predicted binding sites of lncRNA- XLOC_098131 and miR-1180s. (G) Predicted binding sites of JUN and miR-1180s.
Table 5.
Differentially expressed lncRNAs between the daidzein-supplemented group and the daidzein-deficient group.
| Gene_id | Locus | log2(fold_change) | q_value |
|---|---|---|---|
| XLOC_098131 | 8:25819102-25826005 | 2.25348 | 0.002783 |
| XLOC_053963 | 20:5515116-5515913 | 2.49987 | 0.004555 |
| XLOC_101028 | 9:17660403-17663648 | 1.95707 | 0.010438 |
| XLOC_108821 | JH375740.1:3495-5053 | 2.92619 | 0.010736 |
| XLOC_110897 | Z:13526884-13531884 | 1.64843 | 0.012525 |
| XLOC_106453 | AADN03020969.1:48-1144 | 1.98815 | 0.021471 |
| XLOC_060507 | 3:4438760-4440121 | 2.22644 | 0.02505 |
| XLOC_107859 | AADN03026913.1:0-1654 | 1.40834 | 0.029407 |
| XLOC_060807 | 3:44996672-45004147 | 1.27564 | 0.045252 |
| XLOC_107884 | AADN03027032.1:5-1157 | −4.00884 | 0.002783 |
| XLOC_106739 | AADN03022135.1:0-1077 | −3.96327 | 0.002865 |
| XLOC_106700 | AADN03021946.1:8-1557 | −3.12889 | 0.002987 |
| XLOC_107647 | AADN03026044.1:748-1538 | −3.03662 | 0.003083 |
| XLOC_105225 | AADN03015717.1:329-1427 | −2.81266 | 0.003313 |
| XLOC_107752 | AADN03026468.1:23-1490 | −2.25076 | 0.003465 |
| XLOC_106646 | AADN03021718.1:202-1080 | −2.00632 | 0.002567 |
| XLOC_105024 | AADN03014889.1:21-1062 | −1.63664 | 0.002783 |
| XLOC_105701 | AADN03017712.1:5-1287 | −1.80265 | 0.004555 |
| XLOC_031304 | 14:13653237-13661582 | −1.5396 | 0.010736 |
| XLOC_109853 | JH376401.1:5284-11263 | −1.54183 | 0.012525 |
| XLOC_109110 | JH376055.1:0-1722 | −1.40485 | 0.014735 |
| XLOC_106264 | AADN03020084.1:8-995 | −2.81169 | 0.020875 |
| XLOC_109206 | JH376151.1:21542-23697 | −1.84632 | 0.021095 |
| XLOC_107372 | AADN03024860.1:0-1023 | −2.79506 | 0.021293 |
| XLOC_105135 | AADN03015347.1:7-363 | −1.42235 | 0.037575 |
| XLOC_105285 | AADN03015983.1:0-2072 | −1.44937 | 0.043086 |
| XLOC_106988 | AADN03023146.1:0-1058 | −4.39913 | 0.043356 |
| XLOC_107076 | AADN03023608.1:86-1023 | −1.37271 | 0.045232 |
| XLOC_104526 | AADN03012670.1:70-1718 | −1.30948 | 0.042342 |
| XLOC_107295 | AADN03024536.1:0-2094 | −1.11588 | 0.045252 |
| XLOC_109383 | JH376260.1:1543-5603 | −1.08636 | 0.045456 |
Cis-Regulation Analysis of Differential lncRNAs
A screen of all mRNAs within 10 kb from the 31 differentially expressed lncRNAs yielded 392 mRNAs (Table S5). A comparing of these 392 mRNAs with transcriptome mRNA expression data did not result in a significant difference in the expression of these 392 mRNAs between the DS and DAD groups, suggesting that the differentially expressed lncRNAs identified from the comparison of the DAD and DS groups cannot regulate the expression of adjacent mRNAs through cis-regulation.
Potential Binding Sites in the lncRNAs and the mRNAs
To explore the mechanism underlying the action of the differentially expressed lncRNAs and identify the targets, we analyzed the potential binding sites in the lncRNAs and the mRNAs of FOS and JUN, which are key genes that enable the DA-mediated enhancement in the immune response, using mirDB (http://www.mirdb.org/miRDB/). The results showed that lncRNA- XLOC_098131 has two sites for binding to miR-548s (miR-548e-5p), and the seed locations are 484 and 2,722 (Figure 7C). In addition, lncRNA- XLOC_098131 has two binding sites for miR-548s (miR-548ah-5p), and the seed locations are 1,884 and 3,039 (Figure 7D). Furthermore, FOS has a binding site for miR-548s (miR-548ay-3p) located at 1,544 (Figure 7E), and lncRNA- XLOC_098131 has two binding sites for miR-1180s, with seed locations of 961 and 2,501 (Figure 7F). Moreover, JUN has a binding site for miR-1180s located at 1,506 (Figure 7G). These results suggest that lncRNA- XLOC_098131 might regulate the expression of FOS and JUN by competitively binding to miR-548s and miR-1180s.
Characterization of the lncRNA- XLOC_098131 Sequence
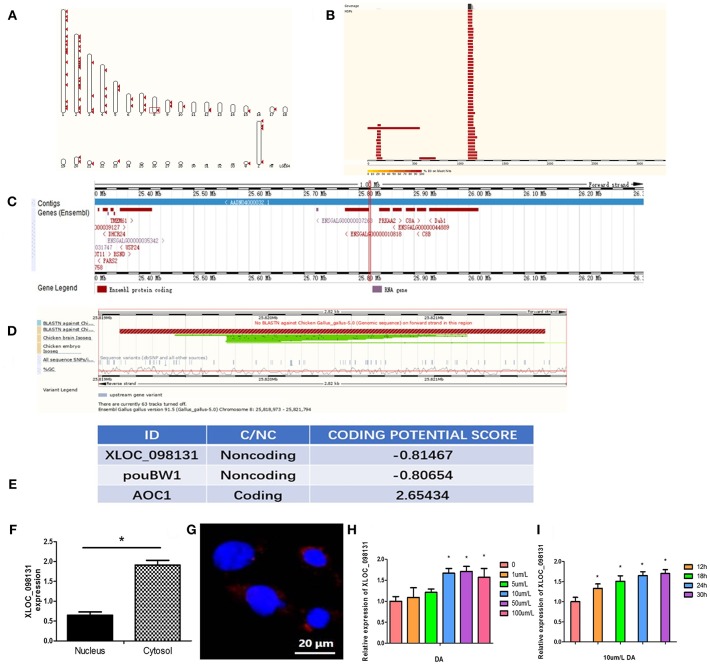
The 5′ and 3′ rapid amplification of cDNA ends (RACE) analyses demonstrated that lncRNA- XLOC_098131 is a transcript consisting of 3,321 nucleotides, and similar to many lncRNAs, this transcript is polyadenylated. The full length of lncRNA- XLOC_098131 is shown in Table S6. The full length of lncRNA- XLOC_098131 was then used in a BLASTN search with the genomic sequences of chicken, human, mouse, pig, cow, and sheep using the online tool Ensemble (http://asia.ensembl.org/) and considering an e-val<0.05. This search yielded more than 100 chicken alignment sequences, one human alignment sequence, one mouse alignment sequence, seven pig alignment sequences, one cow alignment sequence, and four sheep alignment sequences. The results of the first ten comparisons between chickens and other animals are shown in Table 6. For the chicken alignment, the sequence at genomic location 8:25819102–25821666 scored 5,085 with an e-val of 0; the sequence at genomic location 8:25825425–25826005 scored 1,152 with an e-val of 0; and the sequence at genomic location 8:25822459–25822638 scored 357 with an e-val of 1.00E-95. These results are consistent with the transcriptome results, which suggests that lncRNA- XLOC_098131 is located in 8:25819102–25826005. Furthermore, analyses of the high-scoring segment pair (HSP) distribution on the genome (Figure 8A) and HSP distribution on the query sequence (Figure 8B) revealed that lncRNA- XLOC_098131 is located on chromosome 8 in chicken.
Table 6.
BLASTN analysis of the lncRNA- XLOC_098131 sequence with the genomic sequence of multiple species.
| Species | Genomic location | Overlapping gene(s) | Orientation | Query start | Query end | Length | Score | E-val | %ID |
|---|---|---|---|---|---|---|---|---|---|
| Chicken (Gallus gallus) | 8:25819102-25821666 | Reverse | 757 | 3,321 | 2,565 | 5,085 | 0 | 100 | |
| 8:25825425-25826005 | Reverse | 1 | 581 | 581 | 1,152 | 0 | 100 | ||
| 8:25822459-25822638 | Reverse | 580 | 759 | 180 | 357 | 1.00E-95 | 100 | ||
| 6:10061666-10061723 | PRKG1 | Reverse | 1,122 | 1,179 | 58 | 83.8 | 2.00E-13 | 93.1 | |
| 2:102877343-102877419 | Forward | 1,122 | 1,199 | 78 | 83.8 | 2.00E-13 | 89.74 | ||
| 1:13229048-13229148 | RELN | Reverse | 1,122 | 1,222 | 102 | 83.8 | 2.00E-13 | 87.25 | |
| 1:188522666-188522723 | Forward | 1,122 | 1,179 | 58 | 83.8 | 2.00E-13 | 93.1 | ||
| 2:148971284-148971360 | ENSGALG00000046235 | Forward | 1,123 | 1,199 | 77 | 81.8 | 1.00E-12 | 88.31 | |
| 2:60329403-60329449 | Forward | 100 | 146 | 47 | 77.8 | 2.00E-11 | 95.74 | ||
| 2:146079337-146079383 | Forward | 1,122 | 1,168 | 47 | 77.8 | 2.00E-11 | 95.74 | ||
| Human (Homo sapiens) | 16:84600302-84600329 | Forward | 2,952 | 2,979 | 28 | 56 | 0.002 | 100 | |
| Mouse (Mus musculus) | 11:88368731-88368764 | Msi2 | Reverse | 3,005 | 3,038 | 34 | 60 | 2.00E-05 | 97.06 |
| Pig (Sus scrofa) | 10:12597715-12597743 | NVL | Forward | 2,963 | 2,991 | 29 | 50.1 | 0.007 | 96.55 |
| 8:75192172-75192196 | Reverse | 2,952 | 2,976 | 25 | 50.1 | 0.007 | 100 | ||
| 2:5886714-5886738 | Forward | 2,966 | 2,990 | 25 | 50.1 | 0.007 | 100 | ||
| 9:131770635-131770666 | Forward | 2,963 | 2,994 | 32 | 48.1 | 0.028 | 93.75 | ||
| 6:40493835-40493866 | ZNF536 | Forward | 2,963 | 2,994 | 32 | 48.1 | 0.028 | 93.75 | |
| 5:67610724-67610755 | Reverse | 2,963 | 2,994 | 32 | 48.1 | 0.028 | 93.75 | ||
| 1:91969563-91969590 | Forward | 486 | 513 | 28 | 48.1 | 0.028 | 96.43 | ||
| Cow (Bos taurus) | 18:4693512-4693539 | ADAMTS18 | Reverse | 2,963 | 2,990 | 28 | 48.1 | 0.03 | 96.43 |
| Sheep (Ovis aries) | 13:76463289-76463318 | PREX1 | Reverse | 2,950 | 2,979 | 30 | 52 | 0.002 | 96.67 |
| 1:142389415-142389447 | Reverse | 3,008 | 3,040 | 33 | 50.1 | 0.007 | 93.94 | ||
| 3:180456894-180456925 | Reverse | 2,963 | 2,994 | 32 | 48.1 | 0.029 | 93.75 | ||
| 26:36177735-36177762 | Forward | 2,952 | 2,979 | 28 | 48.1 | 0.029 | 96.43 |
Genomic location, location of the hit on the genome; Overlapping gene(s), genes that overlap the genomic location hit; Orientation, strand of the genome the query sequence is found on; Query start and end, positions within the query sequence where the alignment begins and ends (1-based, inclusive); Length, full length of the alignment, including all gaps in either the query or the alignment; Score, BLAST score calculated from the alignment; E-val, probability that the alignment between the query sequence and subject sequence is due to chance; %ID, percentage of the aligned query sequence that is identical to the subject sequence.
Figure 8.
Characterization of the lncRNA-XLOC_098131 sequence. (A) High-scoring segment pair (HSP) is a local alignment with no gaps that achieves one of the highest alignment scores in a given search. It corresponds to the matching region between the query and the database hit sequence. (B) The HSP distribution can be visualized on the query, which is shown as a chain of black and white boxes. Fragments of the query sequence that hit other places in the genome are shown as red boxes. (C) Detailed overview of the region. The gene colors are as follows: gold and red genes: protein coding; gray, blue, and purple genes: non-coding; the red squares indicate the location of lncRNA- XLOC_098131. (D) BLASTN against Chicken Gallus_gallus-5.0 (Genomic sequence), Chicken brain Iso-Seq, and Chicken embryo Iso-Seq. Sequence variants (dbSNP and all other sources): sequence variants from all sources; %GC: percentages of G and C bases in the region. (E) CPC (Coding Potential Calculator) values predicted for lncRNA- XLOC_098131, pouBW1, and AOC1. (F) Expression of lncRNA- XLOC_098131 transcripts in the nuclear and cytosolic of chicken hepatocyte cells. (G) LncRNA- XLOC_098131 FISH of chicken hepatocyte cells. LncRNA- XLOC_098131 is shown in red, and the color blue indicates the cell nucleus. (H) Relative expression of lncRNA- XLOC_098131 in primary chicken hepatocytes after treatment with different doses of daidzein. (I) Relative expression of lncRNA- XLOC_098131 in primary chicken hepatocytes after treatment with daidzein for various times.
As shown in Figure 8C, lncRNA- XLOC_098131 is located on chromosome 8 at position 25819102–25821666 (illustrated by the red line), which is a non-coding area between the two protein-coding genes ENSGALG00000010818 and protein kinase AMP-activated catalytic subunit alpha 2 (PRKAA2) and near the ENSGALG00000044889 gene, which encodes complement C8 alpha chain (C8A), C8B, and DAB1. We performed a further BLASTN analysis, as shown in Figure 8D, and the BLASTN against Chicken Gallus_gallus-5.0 (Genomic sequence) showed no BLASTN pairing with lncRNA- XLOC_098131 and a forward strand orientation in the chicken genome sequence, but BLASTN revealed a high degree of matching with the reverse direction of the chicken genome sequence (lncRNA- XLOC_098131 is a reverse sequence). Furthermore, the BLASTN analysis of lncRNA- XLOC_098131 with the chicken brain Iso-Seq from the European Nucleotide Archive (https://www.ebi.ac.uk/ena) (44) resulted in a high degree of matching, which was slightly lower than that obtained with Chicken Gallus_gallus-5.0 (Genomic sequence); thus, the tissue specificity of lncRNA- XLOC_098131 is low in chickens. The BLASTN analysis of lncRNA- XLOC_098131 with chicken embryo Iso-Seq from the European Nucleotide Archive (https://www.ebi.ac.uk/ena) (44) resulted in a lower degree of matching, showing that the expression level of lncRNA- XLOC_098131 gradually increases with chicken development and highlighting the high temporal specificity of lncRNA- XLOC_098131. The analysis of lncRNA- XLOC_098131 sequence variants from all sources showed that the variability in the sequence of lncRNA- XLOC_098131 is low, indicating that lncRNA- XLOC_098131 has high sequence stability. Furthermore, the percentages of bases G and C in the sequence (%GC) are high, indicating that the sequence of lncRNA- XLOC_098131 has high stability. In addition, consistent with the fact that lncRNA- XLOC_098131 is a non-coding RNA, the Coding Potential Calculator (CPC) computational algorithm predicted that lncRNA- XLOC_098131 has a very low coding potential, similar to pouBW1, which is a well-documented Gallus lncRNA (45) (Figure 8E). These results indicate that lncRNA- XLOC_098131 is a real long non-coding reverse RNA sequence with good stability and low specificity between tissues but with a high temporal specificity and a certain degree of conservation among species.
Cellular Localization of lncRNA- XLOC_098131
To determine the cellular localization of the lncRNA- XLOC_098131 transcript, the nuclear, and cytosolic RNAs from chicken primary hepatocytes were isolated, and the expression of lncRNA- XLOC_098131 transcripts in both subcellular locations was measured. The qRT-PCR data showed that lncRNA- XLOC_098131 transcripts were more highly expressed in the cytosol than in the nucleus (Figure 8F). A FISH analysis of lncRNA- XLOC_098131 in hepatocytes also indicated that lncRNA- XLOC_098131 is mainly located in the cytoplasm (Figure 8G).
DA Treatment Can Upregulate the Expression of lncRNA- XLOC_098131 in vitro
Chicken primary hepatocytes were treated with 0, 1, 5, 10, 50, or 100 μm/L DA for 24 h, and the total RNA from the cells was then extracted for RT-PCR. The 10, 50, and 100 μm/L DA treatment groups exhibited significantly increased expression of lncRNA- XLOC_098131 compared with the control group (Figure 8H). We then examined cultured cells treated with 10 μm/L DA for 0, 12, 18, 24, and 30 h and found that the cells treated with 10 μm/L DA for 12, 18, 24, or 30 h exhibited significantly increased expression of lncRNA- XLOC_098131 compared with the control group (Figure 8I).
Binding Between lncRNA- XLOC_098131 and miR-548s
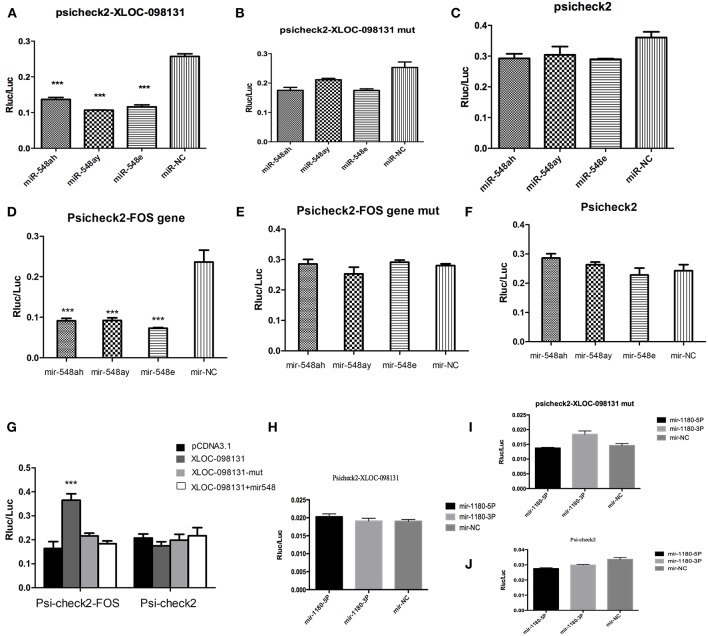
To assess the existence of direct binding between miR-548s and lncRNA- XLOC_098131 at endogenous levels, we constructed luciferase reporters containing lncRNA- XLOC_098131 that contained wild-type (WT) or mutated miR-548s-binding sites. We found that the overexpression of miR-548s reduced the luciferase activities of the WT reporter vector (Figure 9A) but not that of the empty vector (Figure 9C) or that of the mutant reporter vector (Figure 9B). These data demonstrated that the predicted miR-548s could regulate luciferase expression with the 3′UTR of lncRNA- XLOC_098131 (P < 0.05). Mutation of the binding site resulted in the disappearance of this regulation, confirming that miR-548s regulates luciferase through this binding site and that lncRNA- XLOC_098131 might be a target gene of miR-548s.
Figure 9.
Characterization of the lncRNA-XLOC_098131 sequence. (A) Luciferase activity in HEK293FT cells cotransfected with miR-548s and lncRNA-XLOC-098131. (B) Luciferase activity in HEK293FT cells cotransfected with miR-548s and lncRNA-XLOC-098131 mutant transcript. (C) Luciferase activity in HEK293FT cells cotransfected with miR-548s and luciferase reporters containing nothing. (D) Luciferase activity in HEK293FT cells cotransfected with miR-548s and FOS. (E) Luciferase activity in HEK293FT cells cotransfected with miR-548s and FOS mutant transcript. (F) Luciferase activity in HEK293FT cells cotransfected with miR-548s and luciferase reporters containing nothing. (G) Luciferase activity in HEK293FT cells cotransfected with FOS and lncRNA- XLOC_098131, lncRNA- XLOC_098131-mut, lncRNA- XLOC_098131+miR-548s or pCDNA3.1. Luciferase activity in HEK293FT cells cotransfected with Psi-check2 and lncRNA- XLOC_098131, lncRNA- XLOC_098131-mut, lncRNA- XLOC_098131+miR-548s or pCDNA3.1. (H) Luciferase activity in HEK293FT cells cotransfected with miR-1180s and lncRNA-XLOC-098131. (I) Luciferase activity in HEK293FT cells cotransfected with miR-1180s and lncRNA-XLOC-098131 mutant transcript. (J) Luciferase activity in HEK293FT cells cotransfected with miR-1180s and luciferase reporters containing nothing. Data information: The data are presented as the relative ratios of Renilla luciferase activity to firefly luciferase activity.
Binding Between FOS and miR-548s
To validate the hypothesized direct binding between miR-548s and FOS at endogenous levels, we constructed luciferase reporters containing FOS that contained WT or mutated miR-548s-binding sites. We found that miR-548s overexpression reduced the luciferase activities of the WT reporter vector (Figure 9D) but not that of the empty vector (Figure 9F) or that of mutant reporter vector (Figure 9E). These data demonstrated that the predicted miR-548s could regulate luciferase expression with the 3′UTR of FOS (P < 0.05). This regulation disappeared if the binding site was mutated, confirming that miR-548s regulates luciferase through this binding site and that FOS might be a target gene of miR-548s.
LncRNA- XLOC_098131 Upregulates FOS Expression by Serving as a Competitive Endogenous RNA
Because lncRNA- XLOC_098131 shares the regulatory miR-548s with FOS, we questioned whether lncRNA- XLOC_098131 could modulate FOS by regulating miR-548s. As shown in Figure 9G, the overexpression of lncRNA- XLOC_098131 WT, but not that of the mutant or lncRNA- XLOC_098131, increased the FOS transcript level. In addition, the co-transfection of the lncRNA- XLOC_098131 WT overexpression vector, FOS luciferase vector, and sufficient miR-548s resulted in no difference in the luciferase activity of the FOS luciferase vector compared with the control group, and the above treatments had no effect on the psiCHECK-2 luciferase activity.
Binding Between lncRNA- XLOC_098131 and miR-1180s
To validate the existence of direct binding between miR-1180s and lncRNA- XLOC_098131 at endogenous levels, we constructed luciferase reporters containing lncRNA- XLOC_098131 that contained WT or mutated miR-1180s-binding sites. However, compared with the control, the overexpression of miR-1180s could not reduce the luciferase activities of the WT reporter vector (Figure 9H), the mutant reporter vector (Figure 9I) or the empty vector (Figure 9J). This result showed that lncRNA- XLOC_098131 cannot bind to miR-1180s through our predicted binding site. The regulation of the JUN gene by DA treatment is not achieved through the competitive regulation of lncRNA- XLOC_098131, and thus, the mechanism underlying the regulation of JUN expression by DA treatment needs further investigation.
Discussion
Immunoglobulins (Ig), which are key factors in the immune response, prevent infection, can facilitate lysis and activate complements to promote phagocytosis. Previous studies have shown that treating bulls with 100, 200, or 400 ppm DA for 60 days increased their IgA and IgM levels (46), and DA can increase the bovine serum IgE and IgM levels (47). Our results demonstrated that DA supplementation at a dose of 20 mg/kg increased the serum IgA and IgM levels in hens on the 4 and 8th weeks of the experiment, which is consistent with previous research. The peripheral blood lymphocyte content and proliferation ability are important indicators of the body's immune response, and appropriately high levels of lymphocytes in the peripheral blood and a better ability to respond to stimuli are important manifestations of a good immune response (48, 49). Studies investigating lymphocytes following DA treatment have shown that the ability of offspring broilers' B, but not T, lymphocytes to differentiate might be enhanced by adding DA to the breeder's diet at a dose of 20 mg/kg (35). In our study, 20 mg/kg DA increased the number of B lymphocytes in the peripheral blood in hens but had no significant effect on CD3(+) subset of T lymphocytes or on the CD4(+)/CD8(+) T cell ratio. Furthermore, the DA treatment resulted in an increase in the proliferative responses of B cells, as shown by the changes in the LPS SI values obtained in vitro using peripheral blood from hens on the 8th week of the experiment, but had no effect on the proliferative responses of T cells, which is consistent with the previous finding that 0.5 mg/kg DA has no significant effect on T lymphocyte proliferative responses in 5- to 6-week-old boars (50). These results suggest that the effect of DA on lymphocytes is mainly manifested in the regulation of B lymphocytes and that the effect on T lymphocytes is markedly lower. Enhanced B lymphocyte differentiation produces more plasma cells that can secrete more immunoglobulins, which is consistent with the finding that the DA treatment increased the serum IgA and IgM levels in hens. The transcriptome results showed that compared with the DAD group, the DS group exhibited significantly upregulated IRS2 gene expression, which is associated with the inhibition of B lymphocyte apoptosis. Additionally, DA treatment can promote IgA and IgM secretion by regulating the expression of antigen-presenting related genes, such as LOC100859408, ENSGALG00000004772, and LOC768350. Previous studies have found that soy isoflavones, including DA, can regulate the expression of the IRS2 gene, which might affect B lymphocytes (51). Therefore, treatment with 20 mg/kg DA can reduce B lymphocyte apoptosis, promote the differentiation of B lymphocytes into plasma cells that, in turn, secrete more immunoglobulins, promote the presentation of antigens and improve humoral immunity in hens.
Properly elevated levels of neutrophils and mononuclear cells in the blood can increase the body's ability to respond to external stimuli and bacterial infections and enhance immunity (52, 53). Based on our results, the DS group tended to exhibit an increase in the number of neutrophils in the blood of hens on the 4 and 8th weeks of the experiment (P < 0.1), and the number of monocytes in the DS group was significantly higher than that in the DAD group. The RNA-Seq results showed that the DEGs identified from the comparison of two groups such as EGRI1, CAV2, JUN, FOS, SOD3, and CDH2 are involved in the regulation of the interleukin-8 endocytosis viral entry into the host cell, the regulation of Toll-like receptor 21 signaling pathway and MAPK signaling pathway, which can modulate the neutrophils and macrophages. These results show that DA treatment can enhance the body's resistance to viruses and harmful bacteria, enhance the immune response to various stimuli, and improve the body's innate immunity. Previous studies have demonstrated that DA can regulate the content of monocytes and neutrophils and relieve toxic symptoms in mice (54) and that DA can play a positive role in regulating LPS-induced lung injury by regulating the neutrophil content (55).
The KEGG and correlation analyses performed in this study showed that the DS group had an enhanced ability to respond to multiple stimuli compared with the DAD group, and this enhanced ability was regulated by the MAPK signaling pathway, Toll-like pathway and salmonella-related pathway, which can regulate downstream genes to improve humoral and innate immunity, as discussed above. We found that the key genes regulating these processes are JUN, FOS, EGR1, IRS2, CTSK, and CYR61, and the dimer AP-1 composed of JUN and FOS plays a crucial regulatory role in the pathway. AP-1 can regulate the expression of multiple genes to respond to multiple stimuli, including cytokines, growth factors, stress, and bacterial and viral infections. Some studies have found that an appropriately high expression of AP-1 can enhance the body's immune and antibacterial ability and improve its ability to respond to multiple stimuli (56). The AP-1 family of transcription factors can activate Toll-like receptor agonists and positively regulate interleukin-4 (IL-4) to activate macrophages, thereby enhancing antibacterial activity (57). AP-1 plays a key role in regulating immunity as an intermediate regulatory factor; for example, CR3 and Dectin-1 synthesize macrophage cytokines through synergistic activation of the lipid and Syk-JNK-AP-1 pathways (58). Previous studies have shown that multiple plant estrogens can activate AP-1 expression (59), and the treatment of cells with DA significantly increases their expression of the FOS gene (60). DA can also increase FOS gene and protein expression in humans (61). Therefore, in our study, FOS and JUN were identified as the key genes controlling the immunity of hens following DA treatment. The addition of 20 mg/kg DA to the breeder hen diet can upregulate the expression of FOS, JUN, and AP-1 to regulate the Toll-like receptor signaling pathway, MAPK pathway, salmonella infection signal pathway, and related mRNA expression to enhance macrophage activity and increase blood neutrophils and mononuclear cells. This process improves the body's ability to respond to cytokines, growth factors, stress, and bacterial and viral infections and improves the innate immunity.
This study explored the role of lncRNAs in the regulation of key immunity-related genes following DA treatment. We sequenced lncRNAs that showed differential expression between the DA treatment and control groups, identified 1,096 lncRNAs, and found that the frequency of these lncRNA transcripts was mainly concentrated in the 200–5,000 unit range and that the number of transcript exons ranged from 2 to 4, which is consistent with the characteristics of lncRNAs(6). Furthermore, nine and 22 lncRNAs were significantly upregulated and downregulated after DA treatment. We further analyzed the relationship between the differentially expressed lncRNAs and key mRNAs. Previous studies have shown that lncRNAs can influence the expression of adjacent mRNAs that are located within 10 kb of the lncRNAs through cis-regulatory effects; for example, the lncRNA Malat1 plays a cis-regulatory role in the adult mouse (62), and the lncRNA GAPLINC regulates mRNA by cis-regulation in gastric cancer (63). In this study, we selected 31 differentially expressed lncRNAs for the analysis and found that 392 mRNAs are located near these lncRNAs. However, none of these mRNAs exhibited significantly differential expression between the DAD and DS groups, suggesting that the DA treatment could not regulate the expression of mRNAs by the cis-regulatory function of the lncRNAs that were found to be differentially expressed between the two groups. Many studies have found that lncRNAs function as competitive endogenous RNAs to regulate mRNA expression. For example, the lncRNA CCAT1 upregulates the proliferation and invasion of melanoma cells by inhibiting miR-33a (64), the lncRNA XIST regulates AKT expression by competitively binding miR-494 (65), and the lncRNA TUG1 binds to microRNA-9 to regulate the expression of Bcl2l11 (66). By predicting binding sites using an online tool, we found that lncRNA- XLOC_098131, which is the most highly upregulated lncRNA after DA treatment, might regulate FOS expression by competitively binding to miR-548s and regulate JUN expression by competitively binding to miR-1180s. Therefore, lncRNA- XLOC_098131 was selected for further experimentation. First, we also verified that DA treatment can upregulate lncRNA- XLOC_098131 expression in vitro. We then found that lncRNA- XLOC_098131 is a novel transcript located on chromosome 8 in chickens between 25819102 and 25826005, and a BLASTN analysis showed that this fragment is located in a non-coding sequence region, indicating that lncRNA- XLOC_098131 is a non-coding transcript. For further confirmation, we used the CPC computational algorithm and predicted that lncRNA- XLOC_098131 has a very low coding potential, similarly to pouBW1, a well-documented Gallus lncRNA. As several studies have revealed, lncRNAs have low tissue specificity in animals (67), and we found that a BLASTN analysis of lncRNA- XLOC_098131 with chicken brain Iso-Seq resulted in a high degree of matching, which was slightly lower than that obtained from the BLASTN analysis with the chicken genomic sequence. These findings demonstrate the low tissue specificity of lncRNA- XLOC_098131. However, further analysis revealed that lncRNA- XLOC_098131 has low expression in the chicken embryo and that its expression gradually increases with development, highlighting the high temporal specificity of lncRNA- XLOC_098131, and these results are consistent with previous research (68). LncRNAs are known to have high stability and a certain degree of conservation (69). We found that the variability of the sequence of lncRNA- XLOC_098131 is low and that the percentages of the bases G and C in the sequence (%GC) are high, indicating that the sequence of lncRNA- XLOC_098131 exhibits high stability. These results indicate that lncRNA- XLOC_098131 is a real long non-coding reverse RNA sequence with good stability and low specificity between tissues but with a high temporal specificity and a certain degree of conservation among species. LncRNAs can be distributed in the nucleus or cytoplasm, and lncRNAs in the cytoplasm can regulate the expression of mRNAs as ceRNAs (70). In this study, we conducted PCR and FISH experiments to show that lncRNA- XLOC_098131 is mainly located in the cytoplasm, where lncRNAs can bind to miRNAs through their binding sites and thereby eliminate the miRNA-mediated inhibition of mRNAs, which can also bind to miRNAs (71). We successfully demonstrated that both lncRNA- XLOC_098131 and FOS are target genes of miR-548s and that lncRNA- XLOC_098131 can bind to mir-548s through its binding sites to reduce the level of free miR-548s, which in turn reduces the binding of FOS and miR-548s, eliminating the miR-548s-mediated inhibition of FOS and upregulating the expression of FOS. Furthermore, when found at a sufficient high level, the binding of miR-548s to lncRNA- XLOC_098131 did not affect the intracellular free mir-548s content; thus, there was no change in the number of miR-548s that bound to FOS, and the expression of FOS was unchanged. These results confirmed that lncRNA- XLOC_098131 regulates FOS expression by competitively binding to miR-548s. However, subsequent experiments demonstrated that lncRNA- XLOC_098131 cannot regulate JUN gene expression by competitively binding to miR-1180s. LncRNAs can also affect mRNA expression through epigenetic or transcriptional regulation (6), and the molecular mechanism through which lncRNAs regulate JUN expression needs further investigation.
In summary, our study demonstrates that a novel lncRNA activated by DA can stabilize FOS mRNA by serving as a competitive endogenous RNA and can affect AP-1 with JUN to regulate the MAPK signaling, Toll-like receptor signaling, and salmonella signaling pathways in vivo. Dietary supplement DA can improve the numbers of blood neutrophils and mononuclear cells, and improve the innate immunity response. In addition, by increasing the content and differential ability of B lymphocytes and improving antigen presentation and immunoglobulin secretion, DA enhances the body's humoral immunity. In conclusion, our study shows that the novel lncRNA- XLOC_098131 plays a key role in the molecular mechanism underlying the immune function mediated by DA. This study provides new insight into the molecular mechanisms through which DA affects the Toll-like receptor signaling pathway-related immune response and the mechanisms through which lncRNAs regulate mRNA expression. As direct target genes, miR-548s plays important roles in mediating the regulation of FOS by the lncRNA- XLOC_098131 to affect immunity. These data constitute high-quality resources for future genome and functional research.
Ethics Statement
The experimental animal procedures were approved by the China Agricultural University Animal Care and Use Committee (Beijing, China).
Author Contributions
HF designed and performed experiments, analyzed data, and wrote the paper. ZeL, LG, BZ, and ZhL performed some experiments, and analyzed some data. CN, JL, and MY analyzed some transcriptome data. BS, GL, and DT analyzed some data. JG, SY, and YW performed some experiments. YG initiated the study, designed animal experiments, analyzed data, and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor JL for assisting with the transcriptome data analysis in this study. All authors read and approved the final version of the manuscript.
Footnotes
Funding. This study was financially supported by the National Natural Science Foundation of China: 31872375.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2019.00838/full#supplementary-material
References
- 1.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biology. (2003) 4:9. 10.1186/gb-2003-4-9-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. (2010) 19:R162–8. 10.1093/hmg/ddq362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta. (2017) 1864:1887–99. 10.1016/j.bbamcr.2017.08.001 [DOI] [PubMed] [Google Scholar]
- 4.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. (2017) 355:10. 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. In: Kornberg RD, editor. Annual Review of Biochemistry, Vol 81. Annual Review of Biochemistry. Annual Reviews, Palo Alto: (2012). p. 145–66. 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. (2014) 9:1932–56. 10.1002/cmdc.201300534 [DOI] [PubMed] [Google Scholar]
- 7.Mohammad F, Mondal T, Kanduri C. Epigenetics of imprinted long noncoding RNAs. Epigenetics. (2009) 4:277–86. 10.4161/epi.4.5.9242 [DOI] [PubMed] [Google Scholar]
- 8.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. (2010) 143:46–58. 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang CY, Qiu SL, Sun F, Li W, Wang ZQ, Yue B, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. (2017) 410:50–62. 10.1016/j.canlet.2017.09.012 [DOI] [PubMed] [Google Scholar]
- 10.Xie CR, Wang F, Zhang S, Wang FQ, Zheng S, Li Z, et al. Long noncoding RNA HCAL facilitates the growth and metastasis of hepatocellular carcinoma by acting as a ceRNA of LAPTM4B. Mol Ther Nucleic Acids. (2017) 9:440–51. 10.1016/j.omtn.2017.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pang KC, Dinger ME, Mercer TR, Malquori L, Grimmond SM, Chen WS, et al. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J Immunol. (2009) 182:7738–48. 10.4049/jimmunol.0900603 [DOI] [PubMed] [Google Scholar]
- 12.Wang P, Xue YQ, Han YM, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. (2014) 344:310–3. 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]
- 13.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. (2012) 189:2084–8. 10.4049/jimmunol.1200774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui HC, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, et al. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur J Immunol. (2014) 44:2085–95. 10.1002/eji.201344126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. (2013) 341:789–92. 10.1126/science.1240925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu T, Chi HY, Chen JL, Chen C, Huang YY, Xi H, et al. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene. (2017) 631:29–38. 10.1016/j.gene.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 17.He BM, Bai Y, Kang W, Zhang XP, Jiang XJ. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res. (2017) 7:1704–13. [PMC free article] [PubMed] [Google Scholar]
- 18.Lin ZW, Ge JF, Wang Z, Ren JW, Wang XW, Xiong H, et al. Let-7e modulates the inflammatory response in vascular endothelial cells through ceRNA crosstalk. Sci Rep. (2017) 7:12. 10.1038/srep42498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang R, Li P, Zhang GJ, Lu CY, Wang HQ, Zhao GQ. Long non-coding RNA XLOC_008466 functions as an oncogene in human non-small cell lung cancer by targeting miR-874. Cell Physiol Biochem. (2017) 42:126–36. 10.1159/000477121 [DOI] [PubMed] [Google Scholar]
- 20.Xia T, Liao Q, Jiang XM, Shao YF, Xiao BX, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. (2014) 4:7. 10.1038/srep06088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cassidy A. Potential risks and benefits of phytoestrogen-rich diets. Int J Vitam Nutr Res. (2003) 73:120–6. 10.1024/0300-9831.73.2.120 [DOI] [PubMed] [Google Scholar]
- 22.Wang XX, Wu J, Chiba H, Umegaki K, Yamada K, Ishimi Y. Puerariae radix prevents bone loss in ovariectomized mice. J Bone Miner Metab. (2003) 21:268–75. 10.1007/s00774-003-0420-z [DOI] [PubMed] [Google Scholar]
- 23.Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics. (2013) 8:237–45. 10.4161/epi.23790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vitale DC, Piazza C, Melilli B, Drago F, Salomone S. Isoflavones: estrogenic activity, biological effect and bioavailability. Eur J Drug Metabol Pharmacokinet. (2013) 38:15–25. 10.1007/s13318-012-0112-y [DOI] [PubMed] [Google Scholar]
- 25.Masilamani M, Wei J, Sampson HA. Regulation of the immune response by soybean isoflavones. Immunol Res. (2012) 54:95–110. 10.1007/s12026-012-8331-5 [DOI] [PubMed] [Google Scholar]
- 26.Jahromi SR, Arrefhosseini SR, Ghaemi A, Alizadeh A, Tabriz HM, Togha M. Alleviation of experimental allergic encephalomyelitis in C57BL/6 mice by soy daidzein. Iran J Allergy Asthma Immunol. (2014) 13:256–64. 10.1016/j.ijdevneu.2015.04.295 [DOI] [PubMed] [Google Scholar]
- 27.Kao TH, Wu WM, Hung CF, Wu WB, Chen BH. Anti-inflammatory effects of isoflavone powder produced from soybean cake. J Agric Food Chem. (2007) 55:11068–79. 10.1021/jf071851u [DOI] [PubMed] [Google Scholar]
- 28.Tyagi AM, Srivastava K, Sharan K, Yadav D, Maurya R, Singh D. Daidzein prevents the increase in CD4(+)CD28null T cells and B lymphopoesis in ovariectomized mice: a key mechanism for anti-osteoclastogenic effect. PLoS ONE. (2011) 6:e0021216. 10.1371/journal.pone.0021216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TH, Do MH, Oh YL, Cho DW, Kim SH, Kim SY. Dietary fermented soybean suppresses UVB-induced skin inflammation in hairless mice via regulation of the MAPK signaling pathway. J Agric Food Chem. (2014) 62:8962–72. 10.1021/jf5018252 [DOI] [PubMed] [Google Scholar]
- 30.Choi EY, Jin JY, Lee JY, Choi JI, Choi IS, Kim SJ. Anti-inflammatory effects and the underlying mechanisms of action of daidzein in murine macrophages stimulated with Prevotella intermedia lipopolysaccharide. J Periodont Res. (2012) 47:204–11. 10.1111/j.1600-0765.2011.01422.x [DOI] [PubMed] [Google Scholar]
- 31.Zhang XY, Yang XP, Lin YH, Suo MM, Gong L, Chen JZ, et al. Anti-hypertensive effect of Lycium barbarum L. with down-regulated expression of renal endothelial lncRNA sONE in a rat model of salt-sensitive hypertension. Int J Clin Exp Pathol. (2015) 8:6981–7. [PMC free article] [PubMed] [Google Scholar]
- 32.Ma T, Wang R, Zou X. Dioscin inhibits gastric tumor growth through regulating the expression level of IncRNA HOTAIR. BMC Complement Alternat Med. (2016) 16:383. 10.1186/s12906-016-1360-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai Y, Zang XL, Ma JS, Xu GY. Anti-diabetic effect of Portulaca oleracea L. polysaccharideandits mechanism in diabetic rats. Int J Mol Sci. (2016) 17:14. 10.3390/ijms17081201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic hotair. PLoS ONE. (2013) 8:e0070372. 10.1371/journal.pone.0070372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan H, Lv ZP, Gan LP, Guo YM. Transcriptomics-related mechanisms of supplementing laying broiler breeder hens with dietary daidzein to improve the immune function and growth performance of offspring. J Agric Food Chem. (2018) 66:2049–60. 10.1021/acs.jafc.7b06069 [DOI] [PubMed] [Google Scholar]
- 36.Zhang ZH, Li LT, Luo F, Cheng P, Wu F, Wu Z, et al. Rapid and accurate detection of RMP- and INH- resistant Mycobacterium tuberculosis in spinal tuberculosis specimens by CapitalBio (TM) DNA microarray: a prospective validation study. BMC Infect Dis. (2012) 12:7 10.1186/1471-2334-12-303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan Y, Xu H, Leung RK-K. An optimized protocol for generation and analysis of Ion Proton sequencing reads for RNA-Seq. BMC Genomics. (2016) 17:8. 10.1186/s12864-016-2745-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brazma A, Vilo J. Gene expression data analysis. Microbes Infect. (2001) 3:823–9. 10.1016/s1286-4579(01)01440-x [DOI] [PubMed] [Google Scholar]
- 39.Huang YCT, Li ZW, Carter JD, Soukup JM, Schwartz DA, Yang IV. Fine ambient particles induce oxidative stress and metal binding genes in human alveolar macrophages. Am J Respir Cell Mol Biol. (2009) 41:544–52. 10.1165/rcmb.2008-0064OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang P-L, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods. (2012) 9:1069–76. 10.1038/nmeth.2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. (2014) 42:D222–30. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. (2007) 35:W345–9. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun L, Luo HT, Bu DC, Zhao GG, Yu KT, Zhang CH, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. (2013) 41:8. 10.1093/nar/gkt646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. (2005) 6:31. 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei XX, Kang XT, Liu XJ, Jia LJ, Li H, Li ZJ, et al. Identification and SNP association analysis of a novel gene in chicken. Anim Genet. (2016) 47:125–7. 10.1111/age.12387 [DOI] [PubMed] [Google Scholar]
- 46.Zhao XH, Chen ZD, Zhou S, Song XZ, Ouyang KH, Pan K, et al. Effects of daidzein on performance, serum metabolites, nutrient digestibility, and fecal bacterial community in bull calves. Anim Feed Sci Technol. (2017) 225:87–96. 10.1016/j.anifeedsci.2017.01.014 [DOI] [Google Scholar]
- 47.Han DH, Denison MS, Tachibana H, Yamada K. Effects of estrogenic compounds on immunoglobulin production by mouse splenocytes. Biol Pharma Bull. (2002) 25:1263–7. 10.1248/bpb.25.1263 [DOI] [PubMed] [Google Scholar]
- 48.Datta S, Sarvetnick N. Lymphocyte proliferation in immune-mediated diseases. Trends Immunol. (2009) 30:430–8. 10.1016/j.it.2009.06.002 [DOI] [PubMed] [Google Scholar]
- 49.Ryan-Borchers TA, Park JS, Chew BP, McGuire MK, Fournier LR, Beerman KA. Soy isoflavones modulate immune function in healthy postmenopausal women. Am J Clin Nutr. (2006) 83:1118–25. 10.1093/ajcn/83.5.1118 [DOI] [PubMed] [Google Scholar]
- 50.Genlin W, Xiangying Z, Zhaoyu H, Zhaobin L, Weirong L. Effects of daidzein on body weight gain, serum IGF-I level and cellular immune function in intact male piglets. Asian Austr J Anim Sci. (2002) 15:1066–70. 10.5713/ajas.2002.1066 [DOI] [Google Scholar]
- 51.Kwon DY, Hong SM, Ahn IS, Kim MJ, Yang HJ, Park S. Isoflavonoids and peptides from meju, long-term fermented soybeans, increase insulin sensitivity and exert insulinotropic effects in vitro. Nutrition. (2011) 27:244–52. 10.1016/j.nut.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 52.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–5. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- 53.Schildberger A, Rossmanith E, Eichhorn T, Strassl K, Weber V. Monocytes, peripheral blood mononuclear cells, and THP-1 cells exhibit different cytokine expression patterns following stimulation with lipopolysaccharide. Mediat Inflamm. (2013) 10:697972 10.1155/2013/697972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng H, Fu G, Shen J, Shen K, Xu Z, Wang Y, et al. Ameliorative effect of daidzein on cisplatin-induced nephrotoxicity in mice via modulation of inflammation, oxidative stress, and cell death. Oxid Med Cell Longevity. (2017) 2017:3140680. 10.1155/2017/3140680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng G, Sun B, Li T-z. Daidzein attenuates lipopolysaccharide-induced acute lung injury via toll-like receptor 4/NF-kappaB pathway. Int Immunopharmacol. (2015) 26:392–400. 10.1016/j.intimp.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 56.Yu ZB, Huang C, Zhang Q, Feng WH. Porcine reproductive and respiratory syndrome virus (PRRSV) induces IL-12p40 production through JNK-AP-1 and NF-kappa B signaling pathways. Virus Res. (2016) 225:73–81. 10.1016/j.virusres.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 57.Fontana MF, Baccarella A, Kellar D, Oniskey TK, Terinate P, Rosenberg SD, et al. Myeloid expression of the AP-1 transcription factor JUNB modulates outcomes of type 1 and type 2 parasitic infections. Parasite Immunol. (2015) 37:470–8. 10.1111/pim.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang JH, Lin CY, Wu SY, Chen WY, Chu CL, Brown GD, et al. CR3 and dectin-1 collaborate in macrophage cytokine response through association on lipid rafts and activation of Syk-JNK-AP-1 pathway. PLoS Pathog. (2015) 11:26. 10.1371/journal.ppat.1004985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manna SK. Double-edged sword effect of biochanin to inhibit nuclear factor kappaB: suppression of serine/threonine and tyrosine kinases. Biochem Pharmacol. (2012) 83:1383–92. 10.1016/j.bcp.2012.02.011 [DOI] [PubMed] [Google Scholar]
- 60.Strong AL, Ohlstein JF, Jiang Q, Zhang Q, Zheng SL, Boue SM, et al. Novel daidzein analogs enhance osteogenic activity of bone marrow-derived mesenchymal stem cells and adipose-derived stromal/stem cells through estrogen receptor dependent and independent mechanisms. Stem Cell Res Ther. (2014) 5:17. 10.1186/scrt493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park EK, Shin J, Bae EA, Lee YC, Kim DH. Intestinal bacteria activate estrogenic effect of main constituents puerarin and daidzin of Pueraria thunbergiana. Biol Pharma Bull. (2006) 29:2432–5. 10.1248/bpb.29.2432 [DOI] [PubMed] [Google Scholar]
- 62.Zhang B, Arun G, Mao YS, Lazar Z, Hung GN, Bhattacharjee G, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. (2012) 2:111–23. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu Y, Wang JL, Qian J, Kong X, Tang JT, Wang YC, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. (2014) 74:6890–902. 10.1158/0008-5472.can-14-0686 [DOI] [PubMed] [Google Scholar]
- 64.Lv L, Jia J-Q, Chen J. LncRNA CCAT1 upregulates proliferation and invasion in melanoma cells via suppressing miR-33a. Oncol Res. (2017) 26:201–8. 10.3727/096504017x14920318811749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu S, Xie R, Liu X, Shou J, Gu W, Che X. Long coding RNA XIST contributes to neuronal apoptosis through the downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. Int J Mol Sci. (2017) 18:4. 10.3390/ijms18040732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S, Wang M, Yang H, Mao L, He Q, Jin H, et al. LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem Biophys Res Commun. (2017) 485:167–73. 10.1016/j.bbrc.2017.02.043 [DOI] [PubMed] [Google Scholar]
- 67.Liu N, Zhou Q, Qi YH, Wang H, Yang L, Fan QY. Effects of long non-coding RNA H19 and microRNA let7a expression on thyroid cancer prognosis. Exp Mol Pathol. (2017) 103:71–7. 10.1016/j.yexmp.2017.06.004 [DOI] [PubMed] [Google Scholar]
- 68.Gaiti F, Fernandez-Valverde SL, Nakanishi N, Calcino AD, Yanai I, Tanurdzic M, et al. Dynamic and widespread lncRNA expression in a sponge and the origin of animal complexity. Mol Biol Evol. (2015) 32:2367–82. 10.1093/molbev/msv117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. (2009) 136:629–41. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 70.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. (2013) 3:1113–21. 10.1158/2159-8290.cd-13-0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen LP, Wang Q, Liu RX, Chen ZM, Zhang XQ, Zhou PK, et al. LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. (2018) 46:717–29. 10.1093/nar/gkx1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.