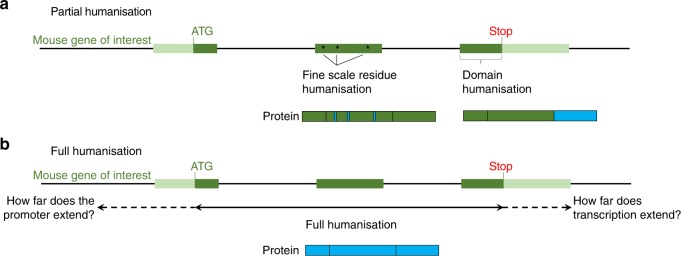
Fig. 2.
How far to humanise. A summary of considerations when deciding on the extent of targeted genomic humanisation for a given gene of interest. Dark green boxes represent exons, lines between exons are introns, light green boxes are UTRs and blue regions represent humanisation. a Partial humanisation typically involves humanising specific residues and/or domains of interest. Fine-scale humanisation of specific amino-acid residues can be performed in isolation if they have known biochemical differences from mouse to human, or if only a small number of residues need to be altered to achieve a human protein sequence. Specific domains or exons can be humanised if, for example, they are known to be critical for human disease. Examples of translated protein products are given. b Full humanisation involves humanising the whole gene, including introns, to attain translation of the full human protein, potentially including human-specific splicing patterns, and for maximum translational potential. 5ʹ and 3ʹ-UTRs, promoters, and other regulatory sequences can be included, on a case-by-case basis, if understanding of gene regulation is the question at hand, if gene clusters are to be humanised or if pathogenic mutations fall within such flanking regions