FIG 4.
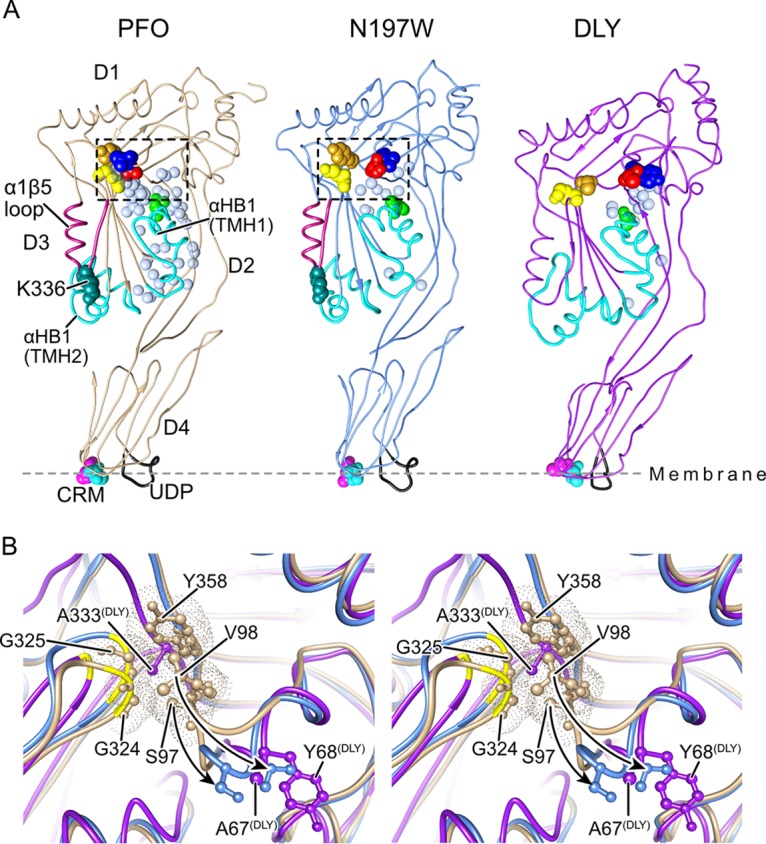
The solution of the crystal structures of DLY and PFON197W reveals differences with PFO at the D3-D1,2 interface. (A) The α-carbon backbone representation of the crystal structures of the previously solved structure of PFO (22) and the structures solved here for PFON197W and DLY. The waters at the D3-D1,2 interface are shown for all three proteins as light blue transparent Van der Waals (VDW) representations. Also shown are the conserved diglycine pairs in D3 (yellow space-filled atoms), the S97 and V98 pair (red and blue space-filled atoms, respectively), Y358 (gold space-filled atoms), the α-helical bundles (αHBs) that ultimately refold to form transmembrane β-hairpins 1 and 2 (cyan), the α1β5 loop (dark red), and the structures of the membrane binding interface: the cholesterol recognition/binding threonine-leucine pair (CRM) and the undecapeptide (UDP, shown in black). (B) Stereo image showing the region boxed in PFO (tan) in panel A overlaid with the same regions in PFON197W (light blue) and DLY (purple), showing the displacement of the D96-Y103 loop (shown by the arrows) in PFON197W and specifically showing the displacement of S97 and V98 from their interaction with the conserved glycine pair (G324, G325), which serves as a flexible linker to allow the rotation of β5 away from β4 (20) and from Y358. The positions of the analogous residues (A67 and Y68) are shown for DLY, and the figure shows that the analogous residue for Y358 in PFO is an alanine in DLY.
