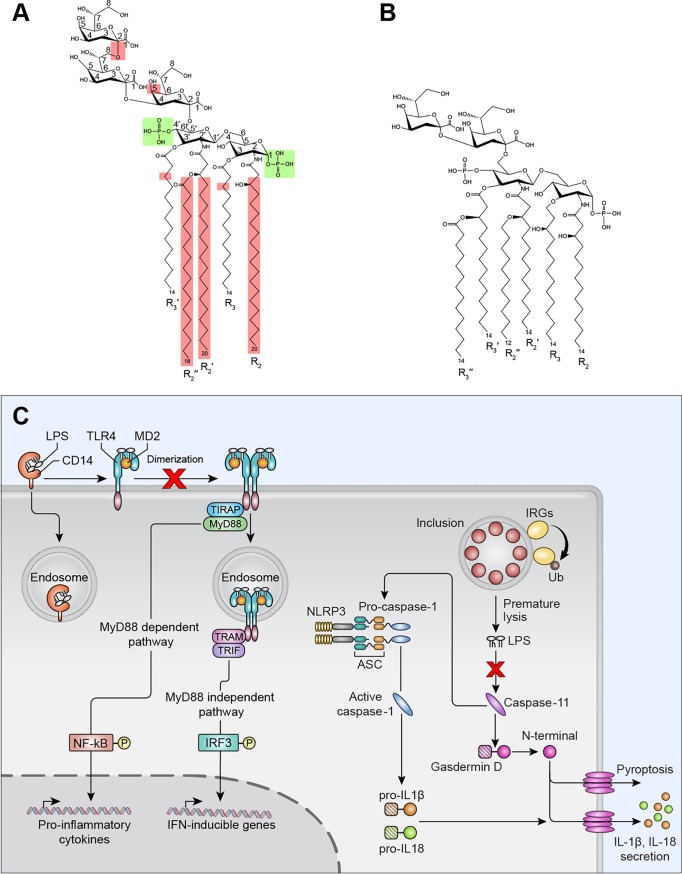
FIG 4.
Structure and functional relationship of C. trachomatis LPS and subversion of canonical and noncanonical inflammatory pathways. (A and B) Structures of chlamydial and E. coli LPS, respectively. C. trachomatis LPS shares the same diphosphorylated glucosamine as E. coli LPS, highlighted in green, an essential structural feature for CD14 binding (25). Structural characteristics of C. trachomatis LPS that differ from E. coli LPS are highlighted in red. (1) A 2-8, 2-4-linked tri-Kdo and the lack of LPS heptosyl transferase 1 (waaC) in the chlamydial genome prevent further glycosylation; thus, chlamydial LPS has a deep rough Re phenotype that avoids PRR recognition. (2) Myristoylated instead of (R)-3-hydroxymyristoyl chains at positions R3′ of Kdo3-lipid IVA, thereby preventing further acylation. (3) Chlamydial LPS amide-linked acyl and 3′ oxyacyl groups (R2, R2′, and R2″) are unusually long, C18–20 compared to C12–14. (C) A model of C. trachomatis LPS subversion of canonical and noncanonical inflammatory pathways.