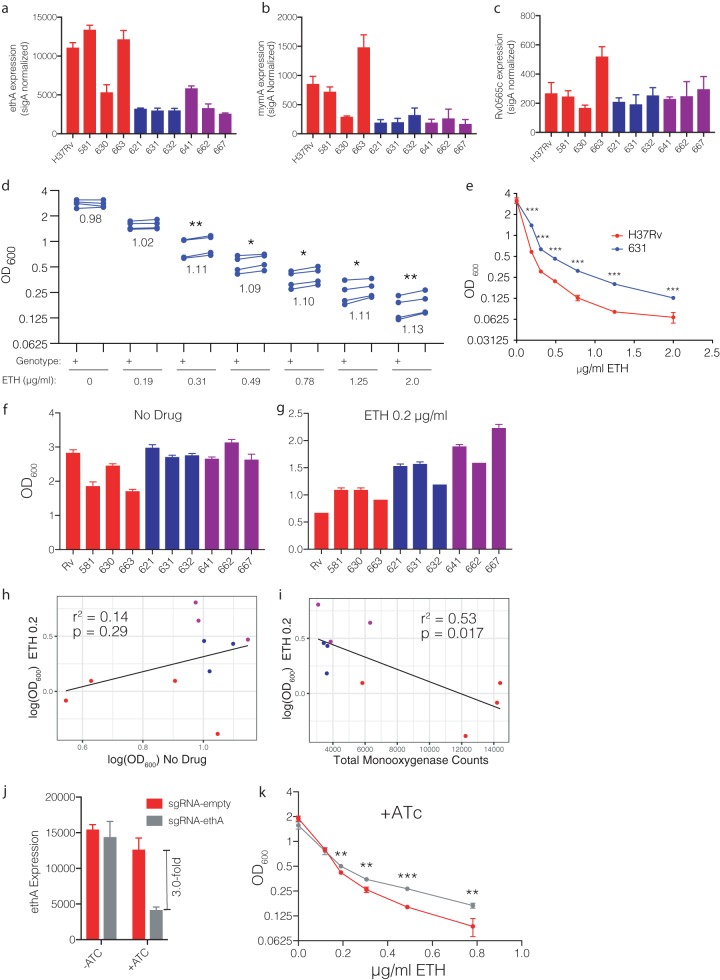
FIG 4.
Monooxygenase expression affects baseline ETH susceptibility. (a to c) Expression of the monooxygenase genes ethA, mymA, and Rv0565c as measured by NanoString and normalized to expression of sigA. Each bar represents the mean normalized expression from three separate cultures, and error bars represent the standard deviation. Colors correspond to lineage designation, with red for lineage 4, blue for lineage 2, and purple for lineage 1. (d) Comparison of the growth of strain 631 (+) and the 631 deletion of Rv0565c (Δ) at various concentrations of ETH. Each point represents the mean growth from three cultures within an experiment with each of four experimental replicates connected by a line. The number below each set of points is the average ratio of deletion over wild type across the 4 experiments. *, P < 0.05; **, P < 0.01, two-tailed paired t test comparing growth at each concentration. (e) Growth of H37Rv and strain 631 across a range of ETH concentrations. Each point represents the mean OD600 from three cultures and is representative of 4 experiments. ***, q < 0.001, two-tailed t tests with false-discovery rate correction for multiple tests. (f and g) Growth of clinical isolates as measured by OD600 after 7 days of growth in either drug-free medium (f) or medium containing 0.2 μg/ml ETH (g). (h and i) Correlation of growth in 0.2 μg/ml ETH with growth in drug-free medium (h) or with the sum of monooxygenase expression across strains (i). The r2 and P values represent the fit of the linear regression plotted on each graph. (j) Knockdown efficiency of ethA in H37Rv as measured by NanoString. Each bar represents the mean ethA expression from three independent cultures normalized to sigA expression with error bars representing the standard deviation. (k) Growth of the H37Rv ethA knockdown strain compared with an empty guide control in the presence of ATc across a range of ETH concentrations. Each point represents the mean from three cultures. **, q < 0.01; ***, q < 0.001, two-tailed t tests with false-discovery rate correction for multiple tests.