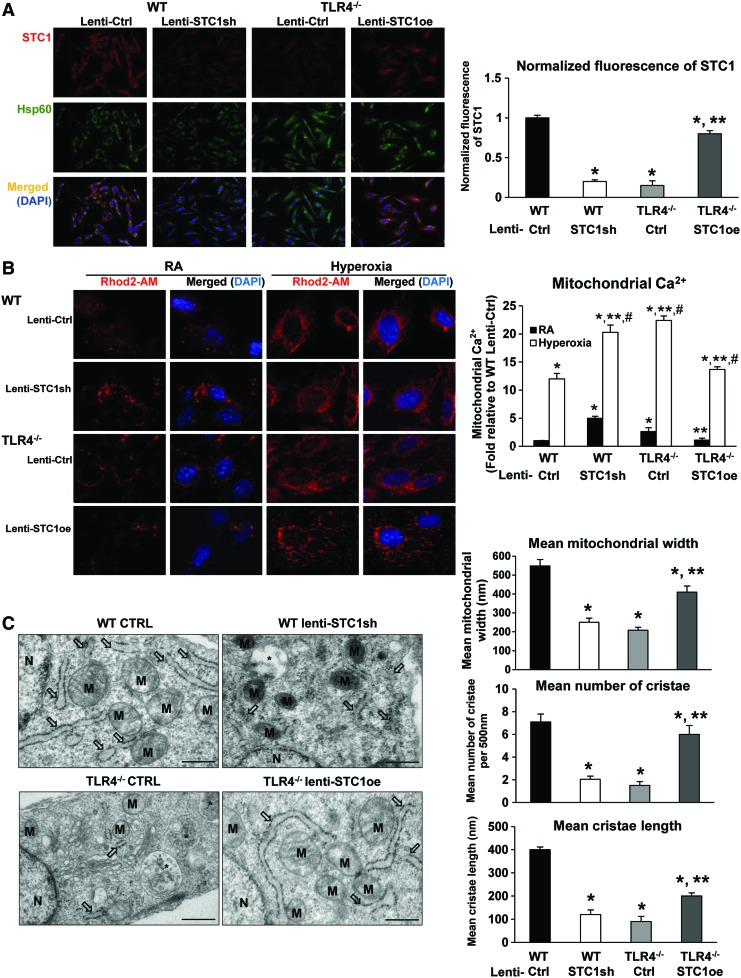
FIG. 7.
STC1 expression correlated with changes in mitochondrial calcium loading and mitochondrial architecture in MLECs. WT MLECs were treated with lentivirus (lenti-Ctrl or lenti-STC1sh) and TLR4−/− MLECs were treated with lentivirus (lenti-Ctrl or STC1 overexpression lenti-STC1oe). (A) Immunostains for STC1 (red) and Hsp60 (mitochondria marker, green) with DAPI-stained nuclei (blue) in MLECs. Left panel shows single immunostained and merged images, original view of all photomicrographs × 400. Right panel shows the normalized fluorescence of STC1 using the ImageJ software. *p < 0.05 vs WT lenti-Ctrl; **p < 0.05 vs TLR4−/− lenti-Ctrl. Data are representative of at least 20 images. (B) MLECs were treated with lentivirus for 48 h followed by incubation with Rhod2-AM for 30 min. Cells were analyzed by fluorescence microscopy after Rhod2-AM staining (red). Nuclei were stained with DAPI (blue). Left panel shows the merged image as an overlay of the DAPI and Rhod2-AM images, original view of all photomicrographs × 1000. Right panel shows the quantification of the mitochondrial Ca2+. *p < 0.05 vs WT lenti-Ctrl RA; **p < 0.05 vs WT lenti-Ctrl hyperoxia; #p < 0.05 vs corresponding RA. Data are representative of at least 20 images. (C) TEM was performed to investigate the ultrastructural differences in MLECs. M, mitochondria; arrows indicate endoplasmic reticulum; *indicates autolysosomes/amphisomes. Scale bar = 500 nm. Right panel: quantification of the mitochondrial width, the number of cristae per 500 nm of mitochondrial length, and the length of cristae mitochondrial ultrastructure organization. *p < 0.05 vs WT lenti-Ctrl; **p < 0.05 vs TLR4−/− lenti-Ctrl. Data are representative of at least 20 cells. Data were analyzed by two-way ANOVA with post hoc Tukey's HSD test calculator for multiple comparisons. TEM, transmission electron microscopy. Color images are available online.