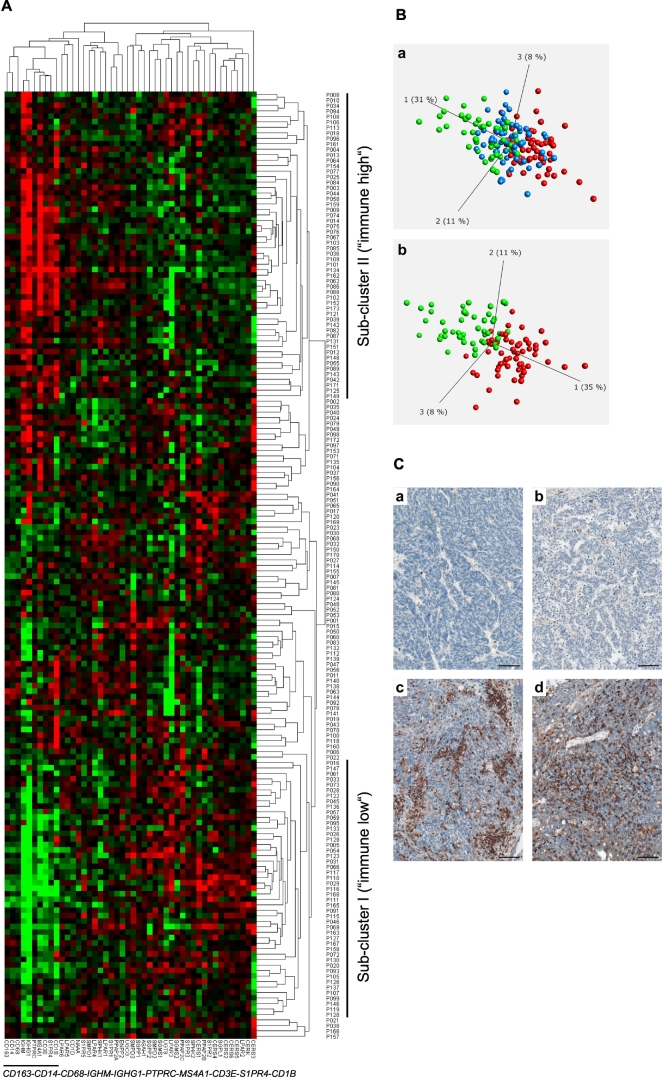
Fig. 4.
Clustering-based patient stratification.
A Pearson uncentered hierarchical clustering was applied across log2 transformed expression values derived from sphingolid/immune-associated multigene signatures-based profiling and across the entire patient population. A result of a clustering run representing a pair of trees is shown for genes and patient samples (Cluster/TreeView programs). Color code: red, high expression and green, low expression according to Cluster/TreeView [91]. The two sub-clusters named as immune low and immune high as well as genes composing the immune-related sub-cluster are indicated.
B A result is shown for PCA that was performed to visualize the data set in a three-dimensional space and identify significantly differential variables between 3 subgroups (a, immune low, mixed, and immune high) and 2 subgroups (b, immune low and immune high) which were obtained on the basis of hierarchical clustering. ANOVA was used for multigroup comparison and t-test for two-group comparison; a p-value cutoff of 0.05 was applied. Color code: green, immune low; blue, mixed; red, immune high.
C Immunohistochemical staining of tissue sections obtained from patients with serous ovarian cancer (clinicopathological characteristics of patient cohort are given in Table S9); Module III in Fig. 1. Images of four representative specimens as shown for the immune low subgroup (a,b) with no/low infiltration of CD45-positive leukocytes and the immune high subgroup (c,d) with massive accumulation of CD45-positive leukocytes within the tumor tissue. Brown color, CD45 staining; blue color, nuclear counterstaining with hematoxylin. Scale bar: 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)