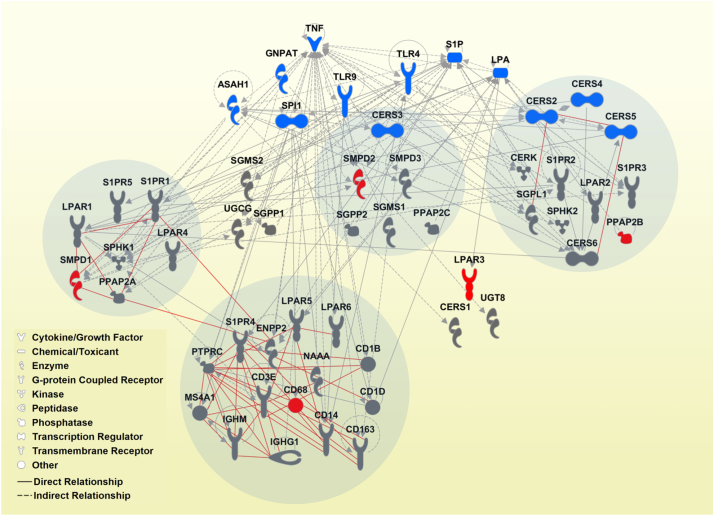
Fig. 12.
Sphingolipid/lysophosphatidate/immune-associated gene network. A reconstructed gene network was created using the IPA software on the basis of genes composing the sphingolipid/lysophosphatidate/immune-associated signature and the top IPA-predicted Upstream Regulators. Grey solid lines display the IPA-identified direct associations between molecules; grey dashed lines display the IPA-identified indirect associations between molecules. Additionally, statistically significant, herein defined biological associations assessed by correlation analysis (correlation coefficient > 0.5, p < .001; Table S2) are displayed by red solid lines. Color code: grey fill, genes composing the multigene signature; red fill, genes with strong prognostic effect (within the top 10 variables of the model Clinics + Sphingo + Immune for OS); blue fill, the top IPA-predicted Upstream Regulators (ASAH1, CERS2, CERS3, CERS4, CERS5 were within the top IPA-predicted Upstream Regulators and are also part of the multigene signature). Grey circles indicate the groups of genes composing the main sub-clusters (> 5 genes, Fig. 3) identified by hierarchical clustering. Insert: the IPA-based description of symbols and relationships. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)