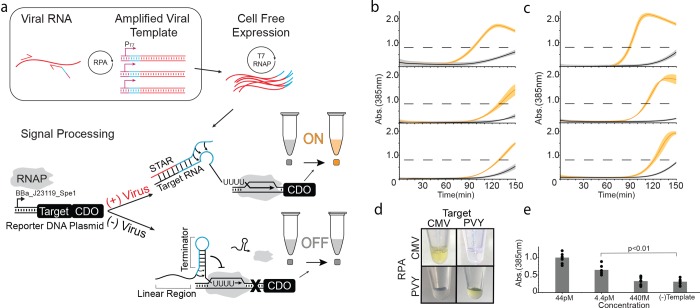
Figure 1.
Basic operation of PLANT-Dx. (a) Schematic overview. Viral RNA is amplified by recombinase polymerase amplification (RPA) into DNA templates that contain a T7 polymerase promoter (purple), a portion of a small transcription activating RNA (STAR) sequence (blue), and a portion of the viral RNA sequence (red). Cell-free expression of these templates produces viral sequence-derived STAR which triggers the production of catechol 2,3-dioxygenase (CDO), which in turn converts catechol into hydroxymuconic semialdehyde, a yellow colored compound that is visible by the eye near a 385 nm absorbance value of 0.8 (dashed line, Figure S3, S4). (b) Demonstrated ability to detect cucumber mosaic virus (CMV) sequences from in vitro transcription (IVT) RNA products (orange) versus control (gray) samples, by 120 min across three different days. (c) Demonstrated ability to detect potato virus Y (PVY) based IVT RNA (orange) versus control (gray) samples by 120 min across three different days. (d) An orthogonality matrix of cell-free reactions challenging RPA products from different IVT sources against different STAR-Target-CDO constructs showing positive results (yellow) only for cognate combinations at 150 min. (e) Serial dilution of CMV IVT RNA was used to determine a limit of detection in between 44 pM and 4.4 pM after 150 min of reaction. (−) Template indicates a control in which no CMV IVT RNA was input into the RPA reaction. p-value comparison was made using a Student’s t test between the data from the 44 pM and (−) Template conditions. Data in (b), (c) represent mean values and error bars represent s.d. of n = 3 technical replicates. Data in (e) represent mean values (bars) of n = 3 biological replicates, each with n = 3 technical replicates (n = 9 total), plotted as individual points.