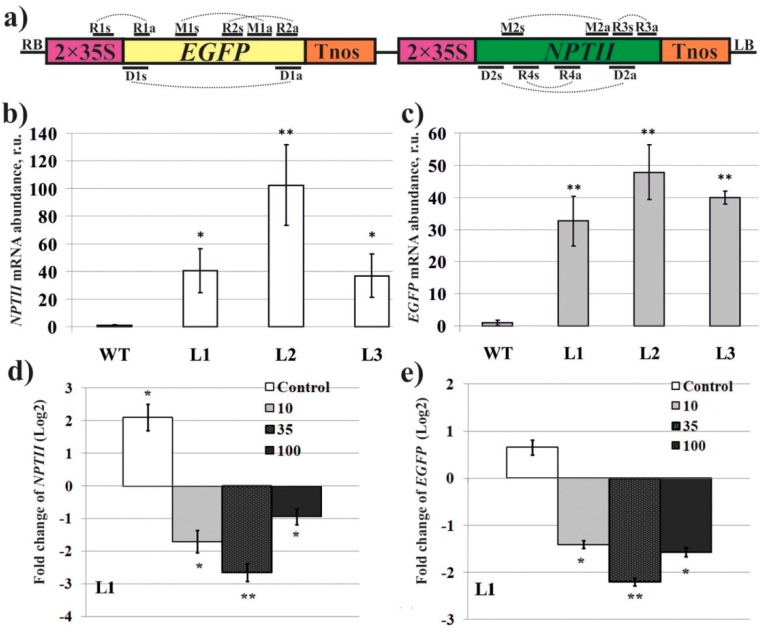
Figure 1.
Characterization of transformed A. thaliana lines, and investigation of the dsRNA concentration effect. (a) Schematic representation of the T-DNA region from the pZP-RCS2-EGFP-NPTII vector [24]. 2x35S—the double 35S promoter of the cauliflower mosaic virus (CaMV); EGFP—the enhanced green fluorescent protein (EGFP) gene; NPTII—the neomycin phosphotransferase II (NPTII) gene; Tnos—nopaline synthase terminator. D1s, D1a, D2s, D2a—primers for dsRNA synthesis; R1s, R1a, R3s, R3a—primers for qRT-PCR estimation of transgene expression after dsRNA treatments; R2s, R2a, R4s, R4a—primers designed to align inside the transgene fragments that have been used for synthesis of the corresponding dsRNAs; M1s, M1a, M2s, M2a—primers for bisulfite DNA sequencing. (b,c) Quantification the NPTII and EGFP mRNAs in the untreated transgenic A. thaliana. RNA was extracted from the wild type (WT) and transformed A. thaliana lines (L1, L2, L3). (d,e) Quantification the NPTII and EGFP mRNAs (log2 fold change) in L1 of A. thaliana in response to external application of NPTII-dsRNA, and EGFP-dsRNA 7 days post-treatment, respectively. 10, 35, 100—the synthesized EGFP-dsRNA and NPTII-dsRNA were diluted in water to concentrations of 0.1, 0.35, and 1 µg/µL (100 µL per plant). Control—sterile filtered water. qRT-PCR data are presented as mean ± SE. *, **—significantly different from the untreated plants at P ≤ 0.05 and 0.01, respectively, according to the Student’s t-test.