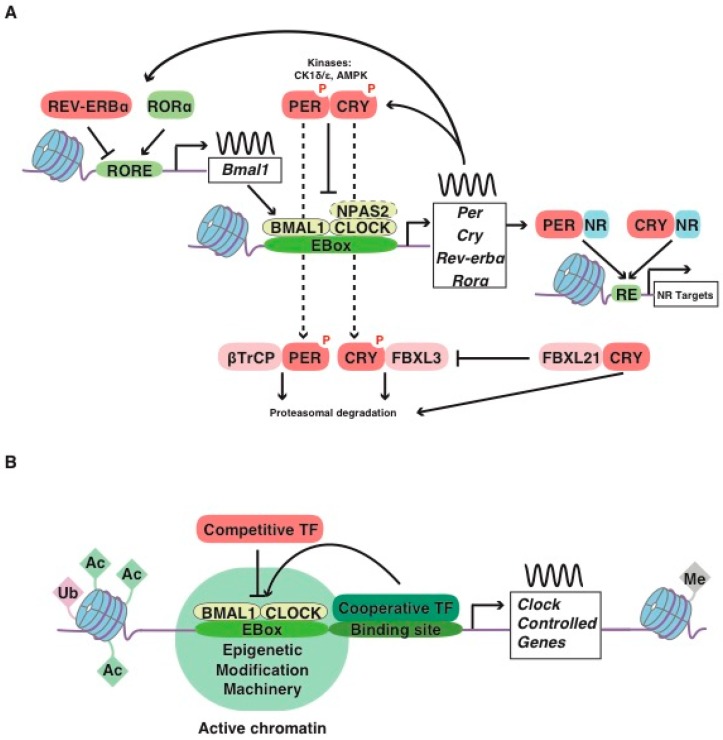
Figure 1.
(A) Core molecular clock network. The mammalian circadian clock consists of transcription/ translation feedback loops in which the positive limb (CLOCK or NPAS2 and BMAL1) heterodimerizes and activates the transcription of downstream genes, including Per, Cry, Rorα and Rev-erbα. The negative limb proteins (PERs and CRYs) multimerize and inhibit CLOCK/BMAL1. The phosphorylation of the repressors mediated by Casein Kinases 1 (CK1ε and CK1δ) and AMPK directs these proteins to ubiquitin-mediated proteasomal degradation (which, for CRY1, is regulated by FBXL protein ratios). In a secondary loop, Bmal1 is regulated by the repressor REV-ERBα and its opposing nuclear receptor RORα, which bind competitively to the shared element RORE, thus repressing or activating the transcription of the Bmal1 gene, respectively (reviewed in [5]). An additional level of circadian regulation exists with CRYs and PERs, which have been reported to bind independently of other core-clock factors to genomic sites enriched with Nuclear Receptor (NR) recognition motifs (response element, RE) [6,7,8,9]. Post-translational modifications of core-clock factors can also regulate transcription (e.g., deacetylation of BMAL1 or PER2 by SIRT1 [10,11]). (B) Regulation of clock-controlled genes. Cell-type and tissue-specific transcription factors bind to specific enhancers and/or respond to specific environmental signals and to chromatin accessibility (regulated by covalent modifications of DNA and histone tails, including histone acetyltransferases such as CBP [12,13], histone deacetylases such as SIRT6 [14], the histone lysine demethylase JARID1a [15], and the histone methyltransferase MLL3 [16], which activate transcription). Transcription factors can compete for CLOCK/BMAL1 binding (e.g., bHLH-ZIP factor USF1 [17], inflammatory factor NF-κB [18], nuclear receptor HNF4A [19], MYC oncogenic factor [20] or through chromatin remodeling (for example the recruitment of the HDAC3 co-repressor by REV-ERBα to regulate BMAL1 binding [21]) or through cooperative effects (lineage-determining transcription factors such as PDX1 [22] or hypoxia-induced HIF1α act synergistically with BMAL1 [23]), thus potentiating or inhibiting the metabolic outputs.