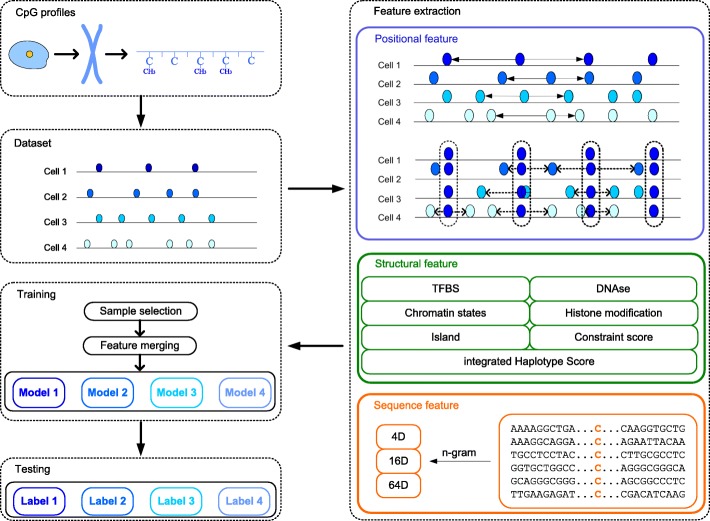
Fig. 1.
The flow chart of LightCpG. CpG profiles are obtained from scTrio-seq. Dataset includes multiple single-cell CpG profiles. Feature extraction: positional feature includes methylation state and the distance between the sites; structural feature includes CpG islands (CGIs) status (CGIs, CGIs shore, CGIs shelf), cis-regulatory elements (TFBS, DNase, chromatin states, histone modification), and DNA properties (integrated haplotype score (iHS), constrain score); sequence feature includes 84 dimension features that are extracted using DNA sequence and n-gram method. Training: LightGBM is used to construct a model for each single-cell CpG data; sample selection is used to reduce the number of samples; feature merging is used to reduce the number of features. Testing: the trained LightCpG model can be used for prediction of the new CpG sites