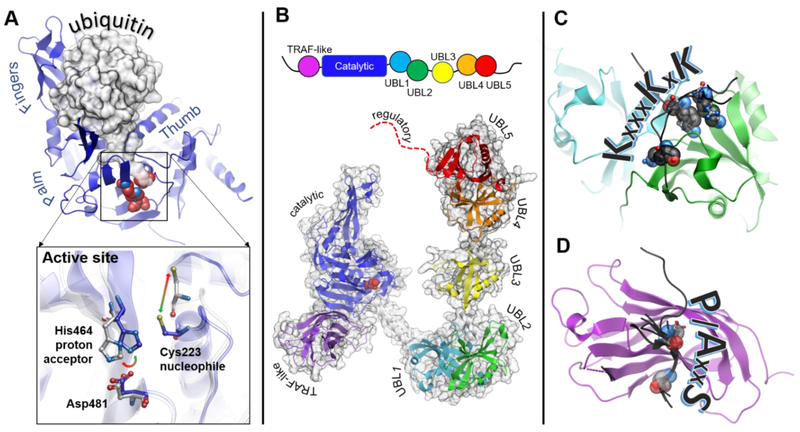
Figure 3. Structural features of USP7.
A. Structure of the catalytic core of USP7 (blue) has a overall right-hand shape formed by Palm, Thumb and Fingers regions. The Fingers grasp ubiquitin molecule, shown as a gray surface. The C-terminal tail of ubiquitin enters the narrow catalytic cleft and extends towards the catalytic triad (shown as red spheres). The inset shows conformational rearrangements of the active site switching between “active” (blue, PDB 1NBF) and “inactive” (grey PDB 1NB8) conformations. B. Schematic representation of the protein domain organization is shown on top. A structural model of the full length USP7 assembled based on overlapping structures of USP7 fragments including PDB IDs 2F1Z, 5FWI and 2YLM. The individual domains are labeled and color-coded: TRAF-like (purple), catalytic (blue), UBL1 (cyan), UBL2 (green), UBL3 (yellow), UBL4 (orange), UBL5 (red), the disordered extreme C-terminus is shown as red dotted line. C. Structure of UBL12 domains (cyan and green, respectively) in complex with its known substrates (black) including DNMT1 (PDB 4YOC), ICP0 (PDB 4WPH) and UHRF1 (PDB 5C6D), The lysine residues of the shared KxxxKxK motif are shown as spheres. The ICP0 peptide has a reversed RxKxxxK motif and binds in the opposite direction. D. Structure of the TRAF-like domain in complex with its known substrates including p53 (PBD 2F1X), HDM2 (PDB 2FOP), HDMX (PDB 3MQR), UbE2E1 (PDB 4JJQ), MCM-BP (PDB 4KG9), vIRF1 (PDB 4YSI), vIRF4 (PDB 2XXN) and EBNA1 (PDB 1YY6). The TRAF-like domain is shown as purple ribbon, and the substrate peptides are black. The P/AxxS motif shows the direction of the peptides. The conserved P/A and S substrate residues are shown as spheres.