Abstract
Human pluripotent stem cells (hPSCs) have extensive applications in fundamental biology, regenerative medicine, disease modelling, and drug discovery/toxicology. Whilst large numbers of cardiomyocytes can be generated from hPSCs, extensive characterization has revealed that they have immature cardiac properties. This has raised potential concerns over their usefulness for many applications and has led to the pursuit of driving maturation of hPSC-cardiomyocytes. Currently, the best approach for driving maturity is the use of tissue engineering to generate highly functional three-dimensional heart tissue. Although we have made significant progress in this area, we have still not generated heart tissue that fully recapitulates all the properties of an adult heart. Deciphering the processes driving cardiomyocyte maturation will be instrumental in uncovering the mechanisms that govern optimal heart function and identifying new therapeutic targets for heart disease.
INTRODUCTION
The maintenance and expansion of embryonic stem cells in a pluripotent state1 have been key to enabling the production of a number of different human cell types, in particular, those that were traditionally hard to expand and culture [e.g., cardiomyocytes (CMs)]. Additionally, the discovery that differentiated adult cells could be reprogrammed back to a pluripotent state using defined factors, first in mouse cells2 and then in human cells,3 now enables the generation of pluripotent stem cells from nearly any patient. These seminal discoveries, together with advanced directed differentiation protocols,4 have led to widespread international use of human pluripotent stem cell (hPSC)-derived cells in both academic and industry led biomedical research.
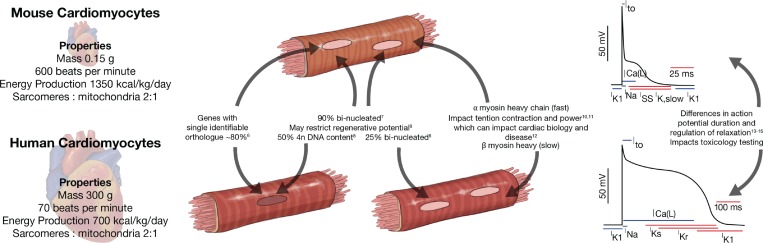
Human cardiomyocytes have many distinct features compared to rodent cardiomyocytes, which have been used as the model system of choice for decades (Fig. 1). To gain a closer representation of human hearts, larger animal models such as rabbits, cats, dogs, sheep, pigs, and monkeys have also been utilized throughout the literature for physiological studies. However, ethical considerations, longer time frames, and higher costs are inhibitory to the widespread use of these larger animal models for routine studies. Therefore, hPSC-derived cardiomyocytes (hPSC-CM) have become an integral part of biological studies and drug discovery in the hope that they may help facilitate translation of research to the clinic.
FIG. 1.
Differences in characteristics of mouse versus human cardiomyocytes.
Many physiological properties such as the size, contraction rate, cardiac output, and metabolism differ between human and mouse hearts.5 Furthermore, around 20% of the protein coding genes in the mouse do not have a single human orthologue.6 These differences give rise to many disparities in the biology of the cardiomyocytes themselves. For example, mouse cardiomyocytes are predominantly bi-nucleated7 compared to human cardiomyocytes being predominantly polypoloid.8 This may have substantial biological significance, as recent studies suggest that that the fraction of binucleated cardiomyocytes has a profound impact on the regenerative potential of the heart.9 Additionally, there are differences in which myosin heavy chain isoforms are predominantly expressed. This impacts how tension and power are generated during contraction, affecting the mechanisms underpinning cardiac disease and function.10–12 Finally, the action potential of mouse and human cardiomyocytes is very different. In fact, this is not just a mouse versus human difference, as the action potential duration tends to scale with the animal size to maintain cardiac output.13–16 Interestingly, many of these differences between mouse and human cardiomyocytes observed in vivo are also observed in CM when mouse pluripotent stem cell and hPSC-CM are compared.17–20 This indicates that while some parameters are dictated by animal physiology, many are also inherently encoded in the genome.
Directed differentiation of hPSC into cardiomyocytes
Decades of research in developmental biology have been essential for the generation of hPSC-CM. This research provided the developmental blueprint to differentiate hPSCs into cardiomyocytes. Originally, non-directed spontaneous differentiation approaches were used (through withdrawal of stem cell maintenance factors). However, these protocols are inconsistent and generate very low percentages of cardiomyocytes. Over the past 15 years, directed differentiation protocols have greatly advanced and have now improved the efficiency of cardiac differentiation to yield greater than 95%. Directed differentiation protocols enable robust and reproducible differentiation and follow known developmental stimuli such as endoderm co-culture,21 growth factor signaling,22–26 and biological pathway manipulation through small molecules.27,28 It is even possible to now purchase hPSC-CM commercially or buy differentiation kits, making hPSC-CM more widely available. These protocols tend to generate predominately ventricular cardiomyocytes although protocols have been recently developed to also efficiently produce nodal29 and atrial cardiomyocyte30,31 sub-types. Therefore, the three major cardiomyocyte sub-types are now readily accessible for biomedical research.
Maturity of hPSC-CM
hPSC-CMs cultured in 2D have many of the key features of their in vivo counterparts. This makes them a very useful and predictive model for many applications including studying hypertrophy, electrophysiology, drug toxicity and discovery, and fundamental biology (reviewed in Ref. 32) However, due to the relative immaturity of 2D hPSC-CM cultures, they have failed as a model for some applications (reviewed in Ref. 33). Responses inconsistent with adult hearts have been observed in studies on: sarcomeric cardiomyopathies such as TITIN mutations associated with dilated cardiomyopathy,34 metabolic syndromes such as mutations in tafazzin,35 cell cycle re-entry where 2D cells respond to mitogens,36 and discovery of bona fide inotropes.37,38 Therefore, induction of hPSC-CM maturation is required to improve modelling capabilities of hPSC-CM.
Why are hPSC-CMs not mature?
Why has maturity similar to that of the adult human heart not yet been achieved? Directed differentiation of hPSC into CM has been very successful, as it builds on decades of research into developmental biology. This research identified the processes that govern the specification and differentiation of early mesoderm and subsequently cardiomyocytes in vivo, which could then be applied to hPSC-CM differentiation. However, the processes known to govern postnatal maturation of the heart are limited and the molecular mechanisms of cardiac maturation remain to be deciphered.
Typical 2D cultures of hPSC-CM cultures fail to reach adult transcriptional maturity even after prolonged culture times (i.e., 1 year).39,40 This indicates that either (1) the development of a fully differentiated adult cardiomyocyte is genetically programmed and may take up to 20 years8 or (2) we have not yet found the key drivers and molecule mechanisms driving adult maturation. Interestingly, some maturation processes such as the cell cycle exit and loss of regeneration capacity occur in a similar time window after birth in both small41,42 and large mammals.43,44 This therefore indicates that some environmental factors are key upstream drivers of maturation and maturation is not solely a genetically timed process. Understanding the environmental drivers of maturation and how they impact cardiomyocytes may therefore help us drive adult maturation in hPSC-CM.
What is maturation?
hPSC-CM maturation is not defined by a single property nor is it driven by a single stimulus. There are a wide array of properties which need to be assessed, covering many aspects of cardiac biology and function (Table I). These require a wide array of assays which need to be performed to assess the different aspects of maturation.39,40,45,46 This is an important point, as adult maturation cannot be stated unless all these properties have been obtained by the hPSC-CM. As Table I highlights, cardiomyocyte properties that change during postnatal heart maturation gear them towards a highly functional and efficient cell. Hence, the key measures that encapsulate multiple aspects of cardiomyocyte maturity are related to their functional properties [Table II (Refs. 8, 9, 36, 39, 40, and 47–64)].
TABLE I.
Properties of immature versus mature cardiomyocytes. F-S: Frank-Starling mechanism where increased sarcomere length leads to increased force of contraction.
| Property | Immature 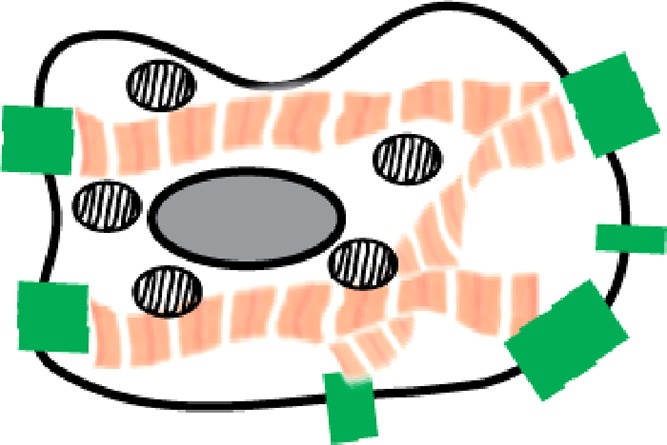
|
Mature 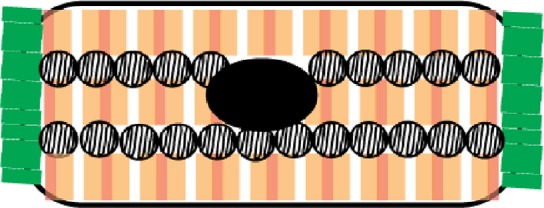
|
Impact with maturation |
|---|---|---|---|
| Sarcomeres | Irregular, 10% volume47 | Organized, 40% volume47 | ↑Force generation |
| 1.8 μm sarcomere spacing48 | 2.2 μm sarcomere spacing48 | ↑Force generation (F-S) | |
| Proteins are fetal isoforms40 | Proteins are adult isoforms40 | Power generation,10–12 ↑stiffness,40,50 signalling51 | |
| Calcium handling | Immature48,52 | Mature48,52 | ↑Calcium amplitude48,52 |
| ↑Force generation | |||
| ↑Faster activation and decay48,52 | |||
| T-tubule system | Poorly developed and organized53 | Highly developed and organized54 | ↑Synchronous and efficient calcium activation throughout the cell54 |
| Ion channel expression | Fetal isoform of INa55 | Adult isoform of INa55 | ↑Upstroke velocity55 |
| Low expression of IK40 | High expression of IK40 | ↓Resting membrane potential56 | |
| Gap junction organization | Circumferential57,58 | Polarized to ends (at intercalated discs)57,58 | ↑Anisotropic conduction velocity |
| ↑Anisotropic force generation | |||
| Metabolism | Glycolytic 10% mitochondria59 | Oxidative phosphorylation 30% mitochondria60,61 | ↑ATP production60,61 |
| ↑Oxygen usage59–61 | |||
| ↑Fatty acids > glucose60,61 | |||
| Cell Cycle | Mitogens drive proliferation36,40 | Mitogens drive hypertrophy62 | ↑Cardiomyocyte size62 |
| ↓Regenerative potential62 | |||
| Nucleus/DNA content | Mono-nucleated monoploid8 | 25% binucleated | ↓Regenerative potential?9 |
| 50% mono-nucleated polyploidy | |||
| 25% mono-nucleated monoploid8 | |||
| ECM binding | β1 integrin collagen I/fibronectin63,64 | Laminin/basement membrane63,64 | ↓Proliferation63,64 |
TABLE II.
Key measurements of maturity. Properties and references are outlined in Table I.
| Contractile force and kinetics |
| Mature cardiomyocytes produce higher forces and have faster upstroke and decay rates. Changes in sarcomere proteins and their organization, calcium handling, t-tubule organization, ion channel expression, and ECM binding all influence contractile force and kinetics. |
| Response to adrenergic stimulation |
| In mature cardiomyocytes, there is a chronotropic, lusitropic, and inotropic response to adrenergic stimulation. Changes in sarcomere proteins and their organization, calcium handling, t-tubule organization, and ion channel expression all influence this response. |
| Increased force-frequency relationship |
| Mature cardiomyocytes increase force as their rate increases (positive staircase). This is heavily influenced by the proteins involved in calcium handling and cellular compartment organization. |
| Conduction velocity |
| Electrical conduction velocity is faster in mature cardiomyocytes. This is regulated by cardiomyocyte coupling via cell-cell connections and gap junctions and by ion channel expression and regulation. Additionally, resting membrane potential of the cardiomyocytes influences this property. |
| Transcriptome |
| The adult cardiomyocyte transcriptome is distinct from an immature cardiomyocyte. There are extensive changes in expression of sarcomeric protein isoforms, metabolic genes, and cell cycle genes during this process.65 The transcriptome can be potentially used as an unbiased holistic measure of maturity40,66 but should be used in combination with functional assays. |
| Metabolism |
| During maturation, there is a switch from glycolysis to fatty acid metabolism. This facilitates a high metabolic capacity and increased mitochondrial biogenesis. |
Bioengineering for maturation
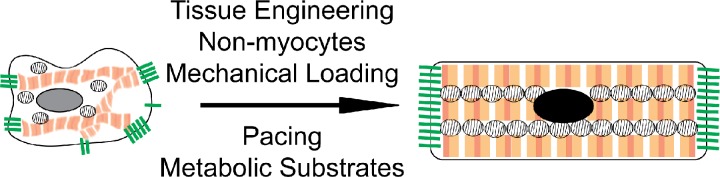
As Table I shows, many functional properties that characterize maturation are the result of the highly controlled organization and compartmentalization of sub-cellular structures. This includes sarcomere alignment, t-tubule organization, formation of sarcoplasmic reticulum adjacent to the t-tubules and sarcomeres, polarized cardiomyocyte-cardiomyocyte junctions to couple both tension and ion flux, organized mitochondria adjacent to the sarcomeres, and cell-cell interactions between different cell types. One way to promote these features is to create a bioengineered environment, allowing the formation of a complex 3D tissue and thus recapitulating in vivo organ-like structures.67 Cells in vivo behave very differently from their in vitro counterparts on stiff, flat, un-patterned 2D substrates.68 Therefore, researchers have used bioengineering to produce cell culture substrates to help mature some properties of cardiomyocytes, including soft hydrogels,69,70 substrate patterning,71–74 or flexible substrates that can deform.74–76 While some of these approaches have generated constructs that achieve high forces of contraction indicative of maturation (up to 10 mN/mm2), a wider array of properties resembling an adult heart have only been shown thus far within 3D engineered heart tissue (EHT) formats.77–79
EHT has been used for decades to create highly functional heart muscle. EHT was originally derived from isolated chicken or neonatal rat heart cells and cultured inside collagen I/Matrigel hydrogels.80,81 Later, fibrin/thrombin based methods have also been used to form EHT.82,83 It is apparent that the most important factor for the extracellular substrate is the ability of the cells to interact with the matrix. By using native ECM such as collagen I/Matrigel/fibrin, the cells can: (1) produce their own matrix which interlocks with these matrices, (2) secrete matrix metalloproteinases to re-arrange the matrix, and (3) bind the matrix for migration, tension generation, and tissue condensation. In addition, it has been shown that co-cultures of different cell types are required in EHT for optimal cardiac function.24,39,84–86 Having a 3D environment based on native matrices enables self-organization of cell-types into in vivo-like cardiac organization,40 thus promoting effective cell-cell communication.
In addition to cellular organization, the EHT approach enables precise mechanical loading of the sarcomeres. In the native heart, most of the mechanical tension is actually imposed on the sarcomeric giant protein TITIN at physiological strains.87 Having a hydrogel full of interconnected cardiac cells is one of the best ways to achieve sarcomeric loading in vitro. This may be an important step for maturation, as tension through cardiomyocyte-substrate binding in stiff 2D environments imposes different biological effects and may actually keep the cardiomyocytes in an immature state63 or impose disease-like states.88 This is also a key consideration for the design of synthetic biomaterials for cardiac tissue engineering, as a stiff polymer or an environment preventing mechanical loading through intercalated discs may be more representative of a 2D rather than a 3D environment.
A variety of different loading regimes have been utilized in the EHT format. Auxotonic mechanical loading, rather than static stretching or phasic stretching, is the most physiological means of EHT mechanical loading and results in enhanced functionality.89–91 This is not surprising as myocardial tissue is inherently designed to respond to mechanical loading for proper heart function. For example, the Frank-Starling mechanism whereby increased preload results in increased force of contraction normalizes beat-to-beat variations in ventricular filling. It is therefore paramount that bioengineering applications carefully consider mechanical loading regimes, as non-physiological loading may have considerable consequences on signaling92 (Table I) and has even been used to model heart disease in EHT.90,93
Overall, the EHT approach has many advantages in creating the most accurate model of heart tissue in vitro. The application of auxotonic mechanical loading and pacing77,79,94 has enabled the production of adult maturity in EHT derived from neonatal rat hearts. Therefore, this approach may also be one of the best methods to achieve adult maturation in hPSC-CM.
Maturation of hPSC-CM
Advanced maturation of hPSC derived-EHT has been achieved by optimizing multi-cellularity and mechanical loading and pacing [Table III (Refs. 20, 23, 39, 40, 45, 46, 65, 93, and 95–107)]. A key finding in multiple studies is that these stimuli drive an increase in EHT function via maturation of the attributes required for optimal cardiac function (see Table I). However, the goal of full adult maturity has not yet been achieved using these approaches, potentially because we do not yet understand all the key drivers and molecular mechanisms of cardiac maturation.
TABLE III.
Properties of EHT benchmarked against 2D culture and adult hearts. Note: Parameters were selected based on analyses of the function (Table II) and those that were measured in the majority of studies. #Tissue produced by the Eschenhagen Lab are more diffuse than other formats, and the functionality seems to be low; but the cardiomyocytes on a per cell basis display a high degree of maturation and are highly functional.
 | ||||
|---|---|---|---|---|
| Approach | Force (mN/mm2) | Isoprenaline force increase (% at EC50 Ca2+) | Mechanical loading regime | |
| Adult heart | 2595 | 20096 | Sarcomeres loaded | |
| Auxotonic | ||||
| Preload: ventricular filling | ||||
| Afterload: systolic blood pressure | ||||
| 2D cardiomyocytes | 0.25–0.5101 | Inconsistent | Dependent on the substrate | |
| Tissue culture plastic has an elastic modulus 100 000 times the heart | ||||
| Most mechanical loading through ECM-integrin rather than sarcomere loading | ||||
| Bursac Lab | Fibrin | 70 | Loaded 3D gel facilitating sarcomeric loading | |
| Zhang et al.45 | Stromal cells | Auxotonic | ||
| Jackman et al.98 | ± Medium convection | −12/+23 | Preload: endogenous cell tension | |
| Shadrin et al.102 | Afterload: undefined–Velcro frame | |||
| Zimmermann Lab | Collagen I | 6.2 | 80–90 | Loaded 3D gel facilitating sarcomeric loading |
| Tiburcy et al.39 | Stromal cells | Auxotonic | ||
| Preload: 10% strain and endogenous cell tension | ||||
| Afterload: controlled by elastic posts | ||||
| Vunjak-Novakovic Lab | Fibrin | 4 | 75 | Loaded 3D gel facilitating sarcomeric loading |
| Ronaldson-Bouchard et al.46 | Stromal cells | Auxotonic | ||
| Pacing | Preload: endogenous cell tension | |||
| Afterload: controlled by elastic posts | ||||
| Sniadecki/Murry Lab | Fibrin | 0.4 | Loaded 3D gel facilitating sarcomeric loading | |
| Leonard et al.93 | Stromal cells | Auxotonic | ||
| Preload: endogenous cell tension | ||||
| Afterload: controlled by elastic posts | ||||
| Murry Lab | Collagen I/Geltrex | 1.3 | Loaded 3D gel facilitating sarcomeric loading | |
| Ruan et al.103 | Stromal cells | Static loading | ||
| Pacing | Preload: endogenous cell tension | |||
| Afterload: dependent on force generation | ||||
| Conklin/Healy Lab | No ECM | 4 | ∼50 | Cardiomycyte/stromal cell mixture adhered to 2D substrate at either end |
| Huebsch et al.104 | Stromal cells | Static loading | ||
| Preload: endogenous cell tension | ||||
| Afterload: dependent on force generation | ||||
| Hudson/Porrello Lab | Collagen I/Matrigel | Loaded 3D gel facilitating sarcomeric loading | ||
| Voges et al.23 | Stromal cells | Auxotonic | ||
| Mills et al.107 | ±Metabolic maturation | 7 | 50 | Preload: endogenous cell tension |
| Mills et al.40 | Afterload: controlled by elastic posts | |||
| Eschanhagen/Hansen Lab | Fibrin | 0.06# | 41 | Loaded 3D gel facilitating sarcomeric loading |
| Schaaf et al.105 | ±Stromal cells | Auxotonic | ||
| Hirt et al.94 | ±Pacing | Preload: endogenous cell tension | ||
| Mannhardt et al.20 | Afterload: controlled by elastic posts | |||
| Ulmer et al.106 | ||||
In addition to the aforementioned stimuli, metabolism has also been recently identified as a major driver of maturation (Table III). In the EHT environment40,106 or even in prolonged 2D culture59,100 (but not to the same extent106), hPSC-CMs increase their capacity for energy production via increased production of mitochondria and in-turn oxidative phosphorylation capacity. This increased respiratory capacity seems to be induced via a contraction-tension based mechanism,106 but further work is required to elucidate the detailed molecular mechanisms involved. Further downstream, PGC-1α has been identified to be likely involved in coordinating these metabolic changes.59,106 While oxidative phosphorylation can increase in the EHT environment with mechanical loading, switching metabolic substrates from glucose/carbohydrates to fatty acids significantly alters cellular metabolism towards oxidative phosphorylation and promotes further maturation.40,100 Exactly how this drives maturation remains to be deciphered, but it is known to be associated with a DNA damage response and the repression of Wnt-β-catenin and yes-associated protein 1/tafazzin signaling.40 Determining the mechanisms behind this maturation process may help us further advance maturation of hPSC-CM.
Similarities to in vivo maturation
In hPSC-CM studies so far, the drivers of maturation are consistent with environmental changes that occur in vivo. In the early postnatal window, there is a shift in many cardiac parameters and recapitulating some of these in hPSC-CM has been shown to result in maturation (Table III). However, there are a considerable number of changes in the postnatal environment (Table IV), and it is still unclear to what extend each of them influences maturation of the heart. Additionally, there are considerable changes in multiple organs during postnatal development, and therefore, inter-organ communication and even interactions between the microbiome and the host108 may be important. The study of these processes not only is pertinent to cardiac maturation but may also be critical of other organ types, as many hPSC-derived cell types are considered “immature.”
TABLE IV.
Factors changing in the postnatal environment (non-exhaustive)
| Oxygen tension |
| Before birth, the arterial oxygen tension is only = 30 mmHg and increases around 3-fold after birth to 100 mm Hg. This has a large impact on metabolism, and the heart transitions from a relatively hypoxic environment where glycolysis is the primary form of energy production to oxidative phosphorylation after birth.97,109 This also creates reactive oxygen species affecting cardiac maturation and additionally has widespread consequences throughout the body.110 |
| Catecholamines (norepinephrine, epinephrine, and dopamine) |
| There is a release of catecholamines following birth to: (1) increase cardiac output, (2) stimulate gluconeogenesis and glycogenolysis in the liver, (3) release free fatty acids, and (4) regulate blood pressure.111 |
| Metabolic substrates |
| Energy production undergoes a switch, as there is a shift from a carbohydrate based to a fatty acid dominated metabolism the newborn starts feeding on breast milk. This induces widespread physiological adaptations such as induction of mitochondrial biogenesis, gluconeogenesis, glycogenolysis, and ketogenesis in the liver to supply other metabolic substrates.112 |
| Serum proteome |
| The serum proteome undergoes major changes after birth.113 Considerable work is required to determine the impact of these changes and which factors are important for cardiac maturation. |
| Cellular composition |
| The cellular composition in the myocardium changes during postnatal maturation. There are varying estimates in the percentage of cells that are cardiomyocytes; however, the general consensus is that the fraction of cardiomyocytes decreases during the maturation period.114–116 The stromal fraction also changes during postnatal maturation;117 in some papers, this has been shown to change from 52:41:6.5 to 52:25:20 endothelial cells:fibroblasts:leukocytes during postnatal maturation and is thus a major change in the fibroblast to leukocyte/macrophage ratio.65 It is still unclear how this influences cardiac maturation and how the different cell populations interact with each other, but there is increasing evidence of complex interplay between the cell types.118 |
Deciphering maturation programs is important for cardiac disease and regeneration
Cardiac dysfunction during disease has many facets and can be attributed to many different processes.119,120 It is well established that the neonatal heart is better adapted to many insults in comparison to the adult heart121 and can functionally regenerate after injury.41–44 Therefore, understanding the maturation process may also lead to the understanding of how the adult heart becomes more susceptible to injury and thus unlock new therapeutic targets for disease. It has been a central dogma for decades that there is a reversion to a fetal phenotype in many disease states, including glycolysis, expression of fetal cardiac proteins, and a growth program (note: hypertrophy not hyperplasia). Reversion of some processes can be cardiac protective, but others can be detrimental. Given the reversion of some properties but not others in disease,65 it is critical that we gain a better understanding of the maturation process so that we can determine the most beneficial adaptions to be exploited as therapeutics.
Application of mature hPSC-CM
Whilst we have not yet produced hPSC-CMs that are equivalent to adult cardiomyocytes, they are still very useful in a wide array of academic and industry based applications, including fundamental science, cell therapy, disease modelling, and drug discovery studies. Furthermore, it should be highlighted that adult hPSC-CM may not be the optimal maturity stage for some applications. For example, for cell therapy applications, engraftment and proliferation following implantation are improved if relatively immature hPSC-CMs are used.122,123 On the other hand, more mature hPSC-CM will be essential for modelling diseases such as Barth syndrome where metabolically mature cardiomyocytes are required.35 Therefore, the bioengineering approach used for different applications may vary considerably (as they currently do), and the optimal bioengineered platform needs to be carefully considered for each specific application.
Scaling of EHT for different applications is also an important consideration. Production and banking of billions of hPSC-CM are now achievable and have already been implemented by some academic labs and within industry. This topic has many aspects requiring consideration and has been extensively reviewed.124 More pertinent to this perspective is the scaling of EHT technologies, especially as culture formats become more and more complex and technically challenging to fabricate. It should also be noted that different applications require different scales. Miniaturization of hPSC-CM36 or EHT40 cultures is required for large-scale drug discovery or biological screening. Conversely, EHT patches for heart regeneration require larger formats.39 When scaling EHT, there are a number of aspects which may require considerable engineering for optimization. Two of the most important considerations are as follows.
Complexity
For all applications, it is essential that the EHT approach used is consistent and reproducible. Therefore, when introducing more complexity into EHT systems, such as pacing, mechanical loading, metabolic substrates, or greater complexities in cell composition, increased quality control is required. When using EHT as a model system, electrically paced EHT formats require that all tissues are paced at the same rate with the same current. In low throughput systems, this is easily managed; however, this becomes increasingly difficult in higher throughput systems where potentially 1000s of miniaturized EHTs are cultured. Furthermore, incorporating more complexity into larger implantable EHT results in additional costs for both the fabrication of the EHT and the additional impact to the Good Manufacturing Practice production pipelines. Overall, additional EHT complexity to enhance maturity requires careful consideration of cost versus benefit when scaling the technology for drug screening or regenerative medicine applications.
Size
The heart is the most metabolically active organ and consumes roughly 20 times its weight in adenosine triphosphate (ATP) daily.125 This has huge consequences on EHT design to ensure sufficient oxygen and nutrient supply. In miniaturized formats where the cell layers are only ∼60 μm (a few cell layers thick) ,40 metabolite or oxygen diffusion is not limited. However, larger formats require strategies to improve supply in order to avoid formation of a necrotic core. A variety of techniques have been successful in achieving this, including having more diffuse muscle bundles,94 incorporating perfused endothelial tubes,126 or having dynamic culture vessels to increase media convection.98
CONCLUSION—HOW CLOSE ARE WE?
The holy-grail of cardiac bioengineering is the production of fully mature adult hPSC-CM, which is also a pursuit for many other organ and cell types.127,128 Driving full adult hPSC-CM maturation will not only result in a better model for many applications but also decipher the poorly understood maturation process of the heart. There has been great progress in this area, and bioengineered models have been instrumental for advanced maturation of many hPSC-CM properties. However, further work is required to progress maturation to an adult state, which will require determination of how the postnatal environment drives maturation. Due to the large number of postnatal physiological changes of the heart, it is likely a multi-faceted approach that mimics mechanical loading, pacing, metabolic substrates, and cellular-composition of the adult heart will be required to drive adult maturation of EHT. How each of these factors influence cardiac maturation and their underpinning molecular mechanisms require detailed follow-up investigations. Understanding of these processes will enable not only the generation of more mature hPSC-CM cultures but also the understanding of how heart biology and function are governed, which in itself may lead to novel therapeutic targets.
ACKNOWLEDGMENTS
J.E.H. was supported by Fellowships from the National Health and Medical Research Council of Australia, the National Heart Foundation of Australia, and the QIMR Berghofer Medical Research Institute.
References
- 1. Thomson J. A. et al. , “ Embryonic stem cell lines derived from human blastocysts,” Science 282, 1145–1147 (1998). 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- 2. Takahashi K. and Yamanaka S., “ Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors,” Cell 126, 663–676 (2006). 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K. et al. , “ Induction of pluripotent stem cells from adult human fibroblasts by defined factors,” Cell 131, 861–872 (2007). 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 4. Tabar V. and Studer L., “ Pluripotent stem cells in regenerative medicine: Challenges and recent progress,” Nat. Rev. Genet. 15, 82–92 (2014). 10.1038/nrg3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kummitha C. M., Kalhan S. C., Saidel G. M., and Lai N., “ Relating tissue/organ energy expenditure to metabolic fluxes in mouse and human: Experimental data integrated with mathematical modeling,” Physiol. Rep. 2, e12159 (2014). 10.14814/phy2.12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Waterston R. H. et al. , “ Initial sequencing and comparative analysis of the mouse genome,” Nature 420, 520–562 (2002). 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- 7. Soonpaa M. H., Kim K. K., Pajak L., Franklin M., and Field L. J., “ Cardiomyocyte DNA synthesis and binucleation during murine development,” Am. J. Physiol. 271, H2183–H2189 (1996). [DOI] [PubMed] [Google Scholar]
- 8. Mollova M. et al. , “ Cardiomyocyte proliferation contributes to heart growth in young humans,” Proc. Natl. Acad. Sci. U. S. A. 110, 1446–1451 (2013). 10.1073/pnas.1214608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patterson M. et al. , “ Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration,” Nat. Genet. 49, 1346–1353 (2017). 10.1038/ng.3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Metzger J. M., Wahr P. A., Michele D. E., Albayya F., and Westfall M. V., “ Effects of myosin heavy chain isoform switching on Ca2+-activated tension development in single adult cardiac myocytes,” Circ. Res. 84, 1310–1317 (1999). 10.1161/01.RES.84.11.1310 [DOI] [PubMed] [Google Scholar]
- 11. Herron T. J., Korte F. S., and McDonald K. S., “ Loaded shortening and power output in cardiac myocytes are dependent on myosin heavy chain isoform expression,” Am. J. Physiol.: Heart Circ. Physiol. 281, H1217–H1222 (2001). 10.1152/ajpheart.2001.281.3.H1217 [DOI] [PubMed] [Google Scholar]
- 12. Davis J. et al. , “ A tension based model distinguishes hypertrophic versus dilated cardiomyopathy,” Cell 165, 1147–1159 (2016). 10.1016/j.cell.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nerbonne J., “ M. Studying cardiac arrhythmias in the mouse—A reasonable model for probing mechanisms?,” Trends Cardiovasc. Med. 14, 83–93 (2004). 10.1016/j.tcm.2003.12.006 [DOI] [PubMed] [Google Scholar]
- 14. Nemtsas P., Wettwer E., Christ T., Weidinger G., and Ravens U., “ Adult zebrafish heart as a model for human heart? An electrophysiological study,” J. Mol. Cell. Cardiol. 48, 161–171 (2010). 10.1016/j.yjmcc.2009.08.034 [DOI] [PubMed] [Google Scholar]
- 15. Kaese S. and Verheule S., “ Cardiac electrophysiology in mice: A matter of size,” Front. Physiol. 3, 345 (2012). 10.3389/fphys.2012.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoekstra M., Mummery C., Wilde A., Bezzina C., and Verkerk A., “ Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias,” Front. Physiol. 3, 346 (2012). 10.3389/fphys.2012.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Janssen B. J., Leenders P. J., and Smits J. F., “ Short-term and long-term blood pressure and heart rate variability in the mouse,” Am. J. Physiol.: Regul., Integr. Comp. Physiol. 278, R215–R225 (2000). 10.1152/ajpregu.2000.278.1.R215 [DOI] [PubMed] [Google Scholar]
- 18. Stoehr A. et al. , “ Spontaneous formation of extensive vessel-like structures in murine engineered heart tissue,” Tissue Eng., Part A 22, 326–335 (2016). 10.1089/ten.tea.2015.0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weinberger F., Mannhardt I., and Eschenhagen T., “ Engineering cardiac muscle tissue: A maturating field of research,” Circ. Res. 120, 1487–1500 (2017). 10.1161/CIRCRESAHA.117.310738 [DOI] [PubMed] [Google Scholar]
- 20. Mannhardt I. et al. , “ Human engineered heart tissue: Analysis of contractile force,” Stem Cell Rep. 7, 29–42 (2016). 10.1016/j.stemcr.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Passier R. et al. , “ Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures,” Stem Cells 23, 772–780 (2005). 10.1634/stemcells.2004-0184 [DOI] [PubMed] [Google Scholar]
- 22. Yang L. et al. , “ Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population,” Nature 453, 524–528 (2008). 10.1038/nature06894 [DOI] [PubMed] [Google Scholar]
- 23. Voges H. K. et al. , “ Development of a human cardiac organoid injury model reveals innate regenerative potential,” Development 144, 1118–1127 (2017). 10.1242/dev.143966 [DOI] [PubMed] [Google Scholar]
- 24. Hudson J. E., Brooke G., Blair C., Wolvetang E., and Cooper-White J. J., “ Development of myocardial constructs using modulus-matched acrylated polypropylene glycol triol substrate and different nonmyocyte cell populations,” Tissue Eng., Part A 17, 2279–2289 (2011). 10.1089/ten.tea.2010.0743 [DOI] [PubMed] [Google Scholar]
- 25. Kattman S. J. et al. , “ Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines,” Cell Stem Cell 8, 228–240 (2011). 10.1016/j.stem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 26. Elliott D. A. et al. , “ NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes,” Nat. Methods 8, 1037–1040 (2011). 10.1038/nmeth.1740 [DOI] [PubMed] [Google Scholar]
- 27. Lian X. et al. , “ Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling,” Proc. Natl. Acad. Sci. U. S. A. 109, E1848–E1857 (2012). 10.1073/pnas.1200250109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burridge P. W. et al. , “ Chemically defined and small molecule-based generation of human cardiomyocytes,” Nat. Methods 11, 855–860 (2014). 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Protze S. I. et al. , “ Sinoatrial node cardiomyocytes derived from human pluripotent cells function as a biological pacemaker,” Nat. Biotechnol. 35, 56–68 (2016). 10.1038/nbt.3745 [DOI] [PubMed] [Google Scholar]
- 30. Cyganek L. et al. , “ Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes,” JCI Insight 3(12), e99941 (2018). 10.1172/jci.insight.99941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee J. H., Protze S. I., Laksman Z., Backx P. H., and Keller G. M., “ Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations,” Cell Stem Cell 21, 179–194.e174 (2017). 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 32. Brandão K. O., Tabel V. A., Atsma D. E., Mummery C. L., and Davis R. P., “ Human pluripotent stem cell models of cardiac disease: From mechanisms to therapies,” Dis. Models Mech. 10, 1039–1059 (2017). 10.1242/dmm.030320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mills R. J., Voges H. K., Porrello E. R., and Hudson J. E., “ Disease modeling and functional screening using engineered heart tissue,” Curr. Opin. Physiol. 1, 80–88 (2018). 10.1016/j.cophys.2017.08.003 [DOI] [Google Scholar]
- 34. Hinson J. T. et al. , “ Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy,” Science 349, 982–986 (2015). 10.1126/science.aaa5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G. et al. , “ Modeling the mitochondrial cardiomyopathy of Barth syndrome with iPSC and heart-on-chip technologies,” Nat. Med. 20, 616–623 (2014). 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Titmarsh D. M. et al. , “ Induction of human iPSC-derived cardiomyocyte proliferation revealed by combinatorial screening in high density microbioreactor arrays,” Sci. Rep. 6, 24637 (2016). 10.1038/srep24637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pointon A. et al. , “ Assessment of cardiomyocyte contraction in human-induced pluripotent stem cell-derived cardiomyocytes,” Toxicol. Sci. 144, 227–237 (2015). 10.1093/toxsci/kfu312 [DOI] [PubMed] [Google Scholar]
- 38. Scott C. W. et al. , “ An impedance-based cellular assay using human iPSC-derived cardiomyocytes to quantify modulators of cardiac contractility,” Toxicol. Sci. 142, 331–338 (2014). 10.1093/toxsci/kfu186 [DOI] [PubMed] [Google Scholar]
- 39. Tiburcy M. et al. , “ Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair,” Circulation 135, 1832–1847 (2017). 10.1161/CIRCULATIONAHA.116.024145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mills R. J. et al. , “ Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest,” Proc. Natl. Acad. Sci. U. S. A. 114, E8372–e8381 (2017). 10.1073/pnas.1707316114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Porrello E. R. et al. , “ Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family,” Proc. Natl. Acad. Sci. U. S. A. 110, 187–192 (2013). 10.1073/pnas.1208863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Porrello E. R. et al. , “ Transient regenerative potential of the neonatal mouse heart,” Science 331, 1078–1080 (2011). 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ye L. et al. , “ Early regenerative capacity in the porcine heart,” Circulation 138(24), 2798–2808 (2018). 10.1161/CIRCULATIONAHA.117.031542 [DOI] [PubMed] [Google Scholar]
- 44. Zhu W. et al. , “ Regenerative potential of neonatal porcine hearts,” Circulation 138(24), 2809–2816 (2018). 10.1161/CIRCULATIONAHA.118.034886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang D. et al. , “ Tissue-engineered cardiac patch for advanced functional maturation of human ESC-derived cardiomyocytes,” Biomaterials 34, 5813–5820 (2013). 10.1016/j.biomaterials.2013.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ronaldson-Bouchard K. et al. , “ Advanced maturation of human cardiac tissue grown from pluripotent stem cells,” Nature 556, 239–243 (2018). 10.1038/s41586-018-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Radisic M. et al. , “ Functional assembly of engineered myocardium by electrical stimulation of cardiac myocytes cultured on scaffolds,” Proc. Natl. Acad. Sci. U. S. A. 101, 18129–18134 (2004). 10.1073/pnas.0407817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lundy S. D., Zhu W. Z., Regnier M., and Laflamme M. A., “ Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells,” Stem Cells Dev. 22, 1991–2002 (2013). 10.1089/scd.2012.0490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van der Velden J. et al. , “ Force production in mechanically isolated cardiac myocytes from human ventricular muscle tissue,” Cardiovasc. Res. 38, 414–423 (1998). 10.1016/S0008-6363(98)00019-4 [DOI] [PubMed] [Google Scholar]
- 50. Neagoe C. et al. , “ Titin isoform switch in ischemic human heart disease,” Circulation 106, 1333–1341 (2002). 10.1161/01.CIR.0000029803.93022.93 [DOI] [PubMed] [Google Scholar]
- 51. Solaro R. J. and Stull J. T., “ Thematic minireview series on signaling in cardiac sarcomeres in health and disease,” J. Biol. Chem. 286, 9895 (2011). 10.1074/jbc.R110.214403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hwang H. S. et al. , “ Comparable calcium handling of human iPSC-derived cardiomyocytes generated by multiple laboratories,” J. Mol. Cell. Cardiol. 85, 79–88 (2015). 10.1016/j.yjmcc.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Parikh S. S. et al. , “ Thyroid and glucocorticoid hormones promote functional T-tubule development in human-induced pluripotent stem cell-derived cardiomyocytes,” Circ. Res. 121, 1323–1330 (2017). 10.1161/CIRCRESAHA.117.311920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brette F. and Orchard C., “ T-tubule function in mammalian cardiac myocytes,” Circ. Res. 92, 1182–1192 (2003). 10.1161/01.RES.0000074908.17214.FD [DOI] [PubMed] [Google Scholar]
- 55. Veerman C. C. et al. , “ Switch from fetal to adult SCN5A isoform in human induced pluripotent stem cell-derived cardiomyocytes unmasks the cellular phenotype of a conduction disease-causing mutation,” J. Am. Heart Assoc. 6(7), e005135 (2017). 10.1161/JAHA.116.005135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Horvath A. et al. , “ Low resting membrane potential and low inward rectifier potassium currents are not inherent features of hiPSC-derived cardiomyocytes,” Stem Cell Rep. 10, 822–833 (2018). 10.1016/j.stemcr.2018.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salameh A. et al. , “ On the role of the gap junction protein Cx43 (GJA1) in human cardiac malformations with Fallot-pathology. A study on paediatric cardiac specimen,” PLoS One 9, e95344 (2014). 10.1371/journal.pone.0095344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vreeker A. et al. , “ Assembly of the cardiac intercalated disk during pre- and postnatal development of the human heart,” PLoS One 9, e94722 (2014). 10.1371/journal.pone.0094722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Birket M. J. et al. , “ PGC-1alpha and reactive oxygen species regulate human embryonic stem cell-derived cardiomyocyte function,” Stem Cell Rep. 1, 560–574 (2013). 10.1016/j.stemcr.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Piquereau J. et al. , “Mitochondrial dynamics adult cardiomyocytes: Which roles a highly specialized cell?” Front. Physiol. 4, 102 (2013). 10.3389/fphys.2013.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Piquereau J. and Ventura-Clapier R., “ Maturation of cardiac energy metabolism during perinatal development,” Front. Physiol. 9, 959 (2018). 10.3389/fphys.2018.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Quaife-Ryan G. A., Sim C. B., Porrello E. R., and Hudson J. E., “ Resetting the epigenome for heart regeneration,” Semin. Cell Dev. Biol. 58, 2–13 (2016). 10.1016/j.semcdb.2015.12.021 [DOI] [PubMed] [Google Scholar]
- 63. Herron T. J. et al. , “ Extracellular matrix-mediated maturation of human pluripotent stem cell-derived cardiac monolayer structure and electrophysiological function,” Circ. Arrhythmia Electrophysiol. 9, e003638 (2016). 10.1161/CIRCEP.113.003638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ieda M. et al. , “ Cardiac fibroblasts regulate myocardial proliferation through β1 integrin signaling,” Dev. Cell 16, 233–244 (2009). 10.1016/j.devcel.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quaife-Ryan G. A. et al. , “ Multicellular transcriptional analysis of mammalian heart regeneration,” Circulation 136, 1123–1139 (2017). 10.1161/CIRCULATIONAHA.117.028252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. DeLaughter D. M. et al. , “ Single-cell resolution of temporal gene expression during heart development,” Dev. Cell 39, 480–490 (2016). 10.1016/j.devcel.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berthiaume F., Maguire T. J., and Yarmush M. L., “ Tissue engineering and regenerative medicine: History, progress, and challenges,” Annu. Rev. Chem. Biomol. Eng. 2, 403–430 (2011). 10.1146/annurev-chembioeng-061010-114257 [DOI] [PubMed] [Google Scholar]
- 68. Humphrey J. D., Dufresne E. R., and Schwartz M. A., “ Mechanotransduction and extracellular matrix homeostasis,” Nat. Rev. Mol. Cell Biol. 15, 802–812 (2014). 10.1038/nrm3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Engler A. J. et al. , “ Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: Scar-like rigidity inhibits beating,” J. Cell Sci. 121, 3794–3802 (2008). 10.1242/jcs.029678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ribeiro A. J. S. et al. , “ Contractility of single cardiomyocytes differentiated from pluripotent stem cells depends on physiological shape and substrate stiffness,” Proc. Natl. Acad. Sci. U. S. A. 112, 12705–12710 (2015). 10.1073/pnas.1508073112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Badie N. and Bursac N., “ Novel micropatterned cardiac cell cultures with realistic ventricular microstructure,” Biophys. J. 96, 3873–3885 (2009). 10.1016/j.bpj.2009.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feinberg A. W. et al. , “ Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture,” Biomaterials 33, 5732–5741 (2012). 10.1016/j.biomaterials.2012.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bray M. A., Sheehy S. P., and Parker K. K., “ Sarcomere alignment is regulated by myocyte shape,” Cell Motility Cytoskeleton 65, 641–651 (2008). 10.1002/cm.20290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Feinberg A. W. et al. , “ Muscular thin films for building actuators and powering devices,” Science 317, 1366–1370 (2007). 10.1126/science.1146885 [DOI] [PubMed] [Google Scholar]
- 75. Oyunbaatar N.-E., Lee D.-H., Patil S. J., Kim E.-S., and Lee D.-W., “ Biomechanical characterization of cardiomyocyte using PDMS pillar with microgrooves,” Sensors 16, 1258 (2016). 10.3390/s16081258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pavesi A. et al. , “ Controlled electromechanical cell stimulation on-a-chip,” Sci. Rep. 5, 11800 (2015). 10.1038/srep11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Godier-Furnémont A. F. G. et al. , “ Physiologic force-frequency in engineered heart muscle by electromechanical stimulation,” Biomaterials 60, 82–91 (2015). 10.1016/j.biomaterials.2015.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tiburcy M. et al. , “ Terminal differentiation, advanced organotypic maturation, and modeling of hypertrophic growth in engineered heart tissue,” Circ. Res. 109, 1105–1114 (2011). 10.1161/CIRCRESAHA.111.251843 [DOI] [PubMed] [Google Scholar]
- 79. Jackman C., Li H., and Bursac N., “ Long-term contractile activity and thyroid hormone supplementation produce engineered rat myocardium with adult-like structure and function,” Acta Biomater. 78, 98–110 (2018). 10.1016/j.actbio.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Eschenhagen T. et al. , “ Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: A new heart muscle model system,” FASEB J. 11, 683–694 (1997). 10.1096/fasebj.11.8.9240969 [DOI] [PubMed] [Google Scholar]
- 81. Zimmermann W. H. et al. , “ Tissue engineering of a differentiated cardiac muscle construct,” Circ. Res. 90, 223–230 (2002). 10.1161/hh0202.103644 [DOI] [PubMed] [Google Scholar]
- 82. Hansen A. et al. , “ Development of a drug screening platform based on engineered heart tissue,” Circ. Res. 107, 35–44 (2010). 10.1161/CIRCRESAHA.109.211458 [DOI] [PubMed] [Google Scholar]
- 83. Bian W., Liau B., Badie N., and Bursac N., “ Mesoscopic hydrogel molding to control the 3D geometry of bioartificial muscle tissues,” Nat. Protoc. 4, 1522–1534 (2009). 10.1038/nprot.2009.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liau B., Jackman C. P., Li Y., and Bursac N., “ Developmental stage-dependent effects of cardiac fibroblasts on function of stem cell-derived engineered cardiac tissues,” Sci. Rep. 7, 42290 (2017). 10.1038/srep42290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Naito H. et al. , “ Optimizing engineered heart tissue for therapeutic applications as surrogate heart muscle,” Circulation 114, I72–I78 (2006). 10.1161/CIRCULATIONAHA.105.001560 [DOI] [PubMed] [Google Scholar]
- 86. Giacomelli E. et al. , “ Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells,” Development 144, 1008–1017 (2017). 10.1242/dev.143438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Granzier H. L. and Irving T. C., “ Passive tension in cardiac muscle: Contribution of collagen, titin, microtubules, and intermediate filaments,” Biophys. J. 68, 1027–1044 (1995). 10.1016/S0006-3495(95)80278-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pandey P. et al. , “ Cardiomyocytes sense matrix rigidity through a combination of muscle and non-muscle myosin contractions,” Dev. Cell 44, 326–336.e323 (2018). 10.1016/j.devcel.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zimmermann W. H. et al. , “ Engineered heart tissue grafts improve systolic and diastolic function in infarcted rat hearts,” Nat. Med. 12, 452–458 (2006). 10.1038/nm1394 [DOI] [PubMed] [Google Scholar]
- 90. Hirt M. N. et al. , “ Increased afterload induces pathological cardiac hypertrophy: A new in vitro model,” Basic Res. Cardiol. 107, 307 (2012). 10.1007/s00395-012-0307-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Boudou T. et al. , “ A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues,” Tissue Eng., Part A 18, 910–919 (2012). 10.1089/ten.tea.2011.0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. LeWinter M. M. and Granzier H., “ Cardiac titin—A multifunctional giant,” Circulation 121, 2137–2145 (2010). 10.1161/CIRCULATIONAHA.109.860171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Leonard A. et al. , “ Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues,” J. Mol. Cell. Cardiol. 118, 147–158 (2018). 10.1016/j.yjmcc.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hirt M. N. et al. , “ Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation,” J. Mol. Cell. Cardiol. 74, 151–161 (2014). 10.1016/j.yjmcc.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 95. Mulieri L. A., Hasenfuss G., Leavitt B., Allen P. D., and Alpert N. R., “ Altered myocardial force-frequency relation in human heart failure,” Circulation 85, 1743–1750 (1992). 10.1161/01.CIR.85.5.1743 [DOI] [PubMed] [Google Scholar]
- 96. Maier L. S. et al. , “ Gingerol, isoproterenol and ouabain normalize impaired post-rest behavior but not force-frequency relation in failing human myocardium,” Cardiovasc. Res. 45, 913–924 (2000). 10.1016/S0008-6363(99)00387-9 [DOI] [PubMed] [Google Scholar]
- 97. Lopaschuk G. D. and Jaswal J. S., “ Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation,” J. Cardiovasc. Pharmacol. 56, 130–140 (2010). 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- 98. Jackman C. P., Carlson A. L., and Bursac N., “ Dynamic culture yields engineered myocardium with near-adult functional output,” Biomaterials 111, 66–79 (2016). 10.1016/j.biomaterials.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kuppusamy K. T. et al. , “ Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes,” Proc. Natl. Acad. Sci. U. S. A. 112, E2785–E2794 (2015). 10.1073/pnas.1424042112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ramachandra C. J. A. et al. , “ Fatty acid metabolism driven mitochondrial bioenergetics promotes advanced developmental phenotypes in human induced pluripotent stem cell derived cardiomyocytes,” Int. J. Cardiol. 272, 288–297 (2018). 10.1016/j.ijcard.2018.08.069 [DOI] [PubMed] [Google Scholar]
- 101. Ribeiro M. C. et al. , “ Functional maturation of human pluripotent stem cell derived cardiomyocytes in vitro–correlation between contraction force and electrophysiology,” Biomaterials 51, 138–150 (2015). 10.1016/j.biomaterials.2015.01.067 [DOI] [PubMed] [Google Scholar]
- 102. Shadrin I. Y. et al. , “ Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues,” Nat. Commun. 8, 1825 (2017). 10.1038/s41467-017-01946-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ruan J. L. et al. , “ Mechanical stress conditioning and electrical stimulation promote contractility and force maturation of induced pluripotent stem cell-derived human cardiac tissue,” Circulation 134, 1557–1567 (2016). 10.1161/CIRCULATIONAHA.114.014998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Huebsch N. et al. , “ Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses,” Sci. Rep. 6, 24726 (2016). 10.1038/srep24726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schaaf S. et al. , “ Human engineered heart tissue as a versatile tool in basic research and preclinical toxicology,” PLoS One 6, e26397 (2011). 10.1371/journal.pone.0026397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ulmer B. M. et al. , “ Contractile work contributes to maturation of energy metabolism in hiPSC-derived cardiomyocytes,” Stem Cell Rep. 10, 834–847 (2018). 10.1016/j.stemcr.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mills R. J., Voges H. K., Porrello E. R., and Hudson J. E., “ Cryoinjury model for tissue injury and repair in bioengineered human striated muscle,” Methods Mol. Biol. 1668, 209–224 (2017). 10.1007/978-1-4939-7283-8 [DOI] [PubMed] [Google Scholar]
- 108. Werder R. B. et al. , “ PGD2/DP2 receptor activation promotes severe viral bronchiolitis by suppressing IFN-lambda production,” Sci. Transl. Med. 10(440), eaao0052 (2018). 10.1126/scitranslmed.aao0052 [DOI] [PubMed] [Google Scholar]
- 109. S. C. Kolwicz, Jr. , Purohit S., and Tian R., “ Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes,” Circ. Res. 113, 603–616 (2013). 10.1161/CIRCRESAHA.113.302095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Puente B. N. et al. , “ The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response,” Cell 157, 565–579 (2014). 10.1016/j.cell.2014.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Morton S. U. and Brodsky D., “ Fetal physiology and the transition to extrauterine life,” Clin. Perinatol. 43, 395–407 (2016). 10.1016/j.clp.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Girard J., Ferre P., Pegorier J. P., and Duee P. H., “ Adaptations of glucose and fatty acid metabolism during perinatal period and suckling-weaning transition,” Physiol. Rev. 72, 507–562 (1992). 10.1152/physrev.1992.72.2.507 [DOI] [PubMed] [Google Scholar]
- 113. Wei L. et al. , “ A comparison of E15.5 fetus and newborn rat serum proteomes,” Proteome Sci. 10, 64 (2012). 10.1186/1477-5956-10-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Banerjee I., Fuseler J. W., Price R. L., Borg T. K., and Baudino T. A., “ Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse,” Am. J. Physiol.: Heart Circ. Physiol. 293, H1883–H1891 (2007). 10.1152/ajpheart.00514.2007 [DOI] [PubMed] [Google Scholar]
- 115. Zhou P. and Pu W. T., “ Recounting cardiac cellular composition,” Circ. Res. 118, 368–370 (2016). 10.1161/CIRCRESAHA.116.308139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bergmann O. et al. , “ Dynamics of cell generation and turnover in the human heart,” Cell 161, 1566–1575 (2015). 10.1016/j.cell.2015.05.026 [DOI] [PubMed] [Google Scholar]
- 117. Pinto A. R. et al. , “ Revisiting cardiac cellular composition,” Circ. Res. 118, 400–409 (2016). 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hulsmans M. et al. , “ Cardiac macrophages promote diastolic dysfunction,” J. Exp. Med. 215, 423–440 (2018). 10.1084/jem.20171274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Burchfield J. S., Xie M., and Hill J. A., “ Pathological ventricular remodeling: Mechanisms: Part 1 of 2,” Circulation 128, 388–400 (2013). 10.1161/CIRCULATIONAHA.113.001878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xie M., Burchfield J. S., and Hill J. A., “ Pathological ventricular remodeling: Therapies: Part 2 of 2,” Circulation 128, 1021–1030 (2013). 10.1161/CIRCULATIONAHA.113.001879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Milerova M. et al. , “ Neonatal cardiac mitochondria and ischemia/reperfusion injury,” Mol. Cell. Biochem. 335, 147–153 (2010). 10.1007/s11010-009-0251-x [DOI] [PubMed] [Google Scholar]
- 122. Liu Y. W. et al. , “ Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates,” Nat. Biotechnol. 36, 597–605 (2018). 10.1038/nbt.4162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Funakoshi S. et al. , “ Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes,” Sci. Rep. 6, 19111 (2016). 10.1038/srep19111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kempf H., Andree B., and Zweigerdt R., “ Large-scale production of human pluripotent stem cell derived cardiomyocytes,” Adv. Drug Delivery Rev. 96, 18–30 (2016). 10.1016/j.addr.2015.11.016 [DOI] [PubMed] [Google Scholar]
- 125. Wang Z. et al. , “ Specific metabolic rates of major organs and tissues across adulthood: Evaluation by mechanistic model of resting energy expenditure,” Am. J. Clin. Nutr. 92, 1369–1377 (2010). 10.3945/ajcn.2010.29885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Vollert I. et al. , “ In vitro perfusion of engineered heart tissue through endothelialized channels,” Tissue Eng., Part A 20, 854–863 (2014). 10.1089/ten.TEA.2013.0214 [DOI] [PubMed] [Google Scholar]
- 127. Mills R. J. et al. , “ Development of a human skeletal micro muscle platform with pacing capabilities,” Biomaterials (published online). 10.1016/j.biomaterials.2018.11.030 [DOI] [PubMed] [Google Scholar]
- 128. Takasato M. et al. , “ Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis,” Nature 526, 564–568 (2015). 10.1038/nature15695 [DOI] [PubMed] [Google Scholar]