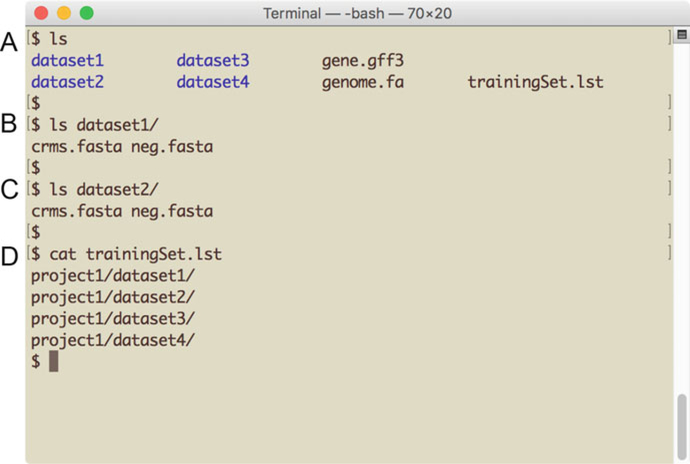
Fig. 2.
Preparing to run SCRMshaw. The screenshot illustrates the directory setup and required user-added files for using SCRMshaw. (a) Directory structure showing the genome file, annotation file, training set list file, and training set subdirectories. (b, c) Each training set subdirectory contains a positive (crms.fasta) and negative (neg.fasta) training data file in the form of a pair of multi-FASTA sequence files. (d) The “trainingSet. lst” file is a text file containing a list of paths to the training set subdirectories