Abstract
Bluetongue virus (BTV) non-structural protein 1 (NS1) regulates viral protein synthesis and exists as tubular and non-tubular forms in infected cells but how tubules assemble and how protein synthesis is regulated are unknown. Here, we report near-atomic resolution structures of two NS1 tubular forms determined by cryo-electron microscopy. The two tubular forms are different helical assemblies of the same NS1 monomer, consisting of an N-terminal foot-, a head- and body domains connected to an extended C-terminal arm, which wraps atop the head domain of another NS1 subunit through hydrophobic interactions. Deletion of the C-terminus prevents tubule formation but not viral replication, suggesting an active non-tubular form. Two zinc finger-like motifs are present in each NS1 monomer and tubules are disrupted by divalent cation chelation and restored by cation addition, including Zn2+, suggesting a regulatory role of divalent cations in tubule formation. In vitro luciferase assays show that the NS1 non-tubular form upregulates BTV mRNA translation, while zinc-finger disruption decreases viral mRNA translation, tubule formation and virus replication, confirming a functional role. Thus, the non-tubular form of NS1 is sufficient for viral protein synthesis and infectious virus replication and the regulatory mechanism involved operates through divalent cation-dependent conversion between the non-tubular and tubular forms.
Introduction
Viruses commonly hijack cellular translational machineries to synthesize viral proteins from their mRNAs using diverse strategies1. In addition, some viruses, exemplified by members of the Reoviridae family, also enhance their gene expression using virally encoded proteins1–4. Among them, rotaviruses and orbiviruses [e.g., bluetongue virus (BTV)] use non-structural (NS) proteins for this purpose. Similar to cellular mRNAs, the 5’ termini of mRNAs of these viruses are capped by viral encoded structural proteins. However, unlike cellular mRNAs and most eukaryotic viruses, the 3’ termini of these viral mRNAs lack a poly(A) tail, which is responsible for enhancing the rate of translation initiation and the stability of RNA. Therefore, in order to specifically upregulate protein synthesis from viral mRNA, viral proteins must bind viral mRNAs selectively over poly(A)-tailed cellular mRNAs. To accomplish this, BTV uses a viral encoded protein, NS1, a 64 kDa protein, which does not bind any poly(A)-tailed mRNA, but has specific binding affinity for the 3’ terminal sequences of BTV mRNAs to upregulates viral protein synthesis4. In rotavirus, the non-structural viral protein NSP3 substitutes the cellular poly(A) binding protein and binds to the viral mRNAs via 3′ terminal sequences2. While biochemical studies and structural studies of NSP3 fragments have yielded some understanding about how this protein may enhance viral mRNA synthesis, high-resolution structures for full-length rotavirus NSP3 and BTV NS1, as well as a detailed molecular understanding of how these viral proteins function, are still lacking3,5–8.
BTV is transmitted by biting midges of the Culicoides genus to ruminants, and is endemic worldwide. BTV infection in sheep and cattle often causes high morbidity and mortality with significant economic consequences. BTV genome consists of 10 segments (S1-S10) of double-stranded (ds) RNA, which encode seven structural proteins and four non-structural proteins (NS1-NS4). NS1 is an early protein expressed in infected host cells at a high level. Within the host cytoplasm, multiple copies of NS1 rapidly assemble in tubular form, a hallmark of orbivirus infections. NS1 tubules can either be purified from BTV infected cells or assembled from recombinant proteins9,10 for structural determination by cryo-electron microscopy (cryoEM) with helical reconstruction. A low-resolution (40 Å) three-dimensional (3D) reconstruction of NS1 tubules generated by recombinant NS1, was previously reported in 19929. However, due to technical limitations, efforts to sort out multiple helical forms in order to improve the resolution of NS1 helical reconstruction have been unsuccessful for the last three decades. As such, how NS1 assemble into helical tubules and how such assemblies participate in BTV gene expression, remain unclear.
Here, we report the near-atomic resolution structures of two NS1 tubular forms obtained by cryoEM. The atomic model shows that each NS1 monomer contains two metal binding, zinc-finger-like motifs and an extended C-terminal arm, which interacts with neighboring subunits to form tubules with variable diameters and helical configurations. The structure rationalizes many of the previous observations attributed to NS1 and suggests how tubules may form from a soluble pool of functional intermediates through coordination of zinc or other metal cations. Further, structure-based mutagenesis of NS1, combined with reverse genetics, allowed us to determine that the non-tubular form of NS1 is also functional and to identify the critical residues involved in viral protein translation and replication, which may be shared by other members of the orbivirus family.
Results
Structure determination of NS1 tubules and atomic modeling
A challenge in the structural study of BTV NS1 protein is its variable tubular forms as recognized nearly 30 years ago9. Such variability makes indexing of the helical parameters difficult and has hitherto prevented achieving high-resolution 3D structures. As technology availed, here we have improved the resolution of the structure (see details in Methods), and determined the multiple helical configurations of the tubules based on our film data but were unable to reach a resolution that is sufficient for atomic modeling. NS1 tubules vary in two major helical settings: subunits per turn (19.xx, 20.xx, 21.xx, 22.xx; where xx means not fixed) and helical start number (1–3 helices). The diameter of the tubule increases as the integer part of the subunits per turn number increases, ranging from 500 Å to 523 Å, 547 Å, 580 Å for 19.2, 20.2, 21.2 and 22.2 subunits per turn, respectively. The single (1-start) helices are always y+1/5 (y in [19.22]) subunits per turn, 2-start y+4/7 subunits per turn and 3-start y+1 subunits per turn. Therefore, we observed, by combination, 12 (4×3) different helical forms, though we do not rule out the existence of other forms of undetectable populations.
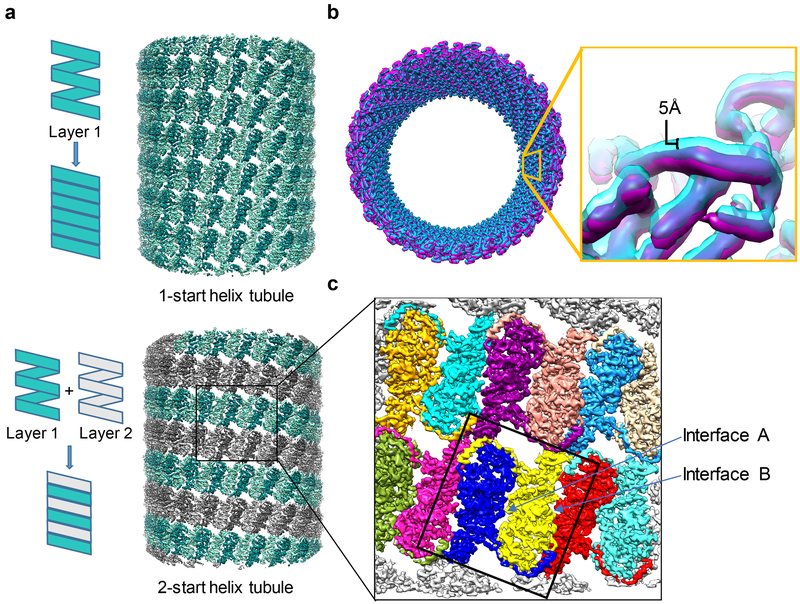
We focused on the two most abundant classes of tubules with the following helical configurations: 20.2 subunits per turn, 1-start helical tubule (17.5%) and 20.58 subunits per turn, 2-start helical tubule (26.8%) (Fig. 1a). The tubule diameter increases from ~523 Å in the 1-start tubule, to ~526 Å in the 2-start tubule. The tubules that belong to the same helical configuration appear to “breathe” due to thermal motions – for four different classes of 3D classification, the radii of 20.58 tubules vary by up to ~5 Å (Fig. 1b). Such variability has necessitated exhaustive computational classification to reduce the number of particles in each homogeneous structural class. Thus, although meaningful signal can be recognized to 3.8 Å in the power spectrum of electron-counting cryoEM images, the resolution of the 3D reconstruction (Fig. 1c) from best class, obtained by merging 4,517 particles, is limited to ~4.0 Å as judged from the structural features (Fig. 2a) and Fourier shell correlation analysis (Fig. S1).
Figure 1: NS1 tubules are dynamic.
(a) Schematic (left) and reconstructions (right) of a 2-start NS1 tubule and a 1-start NS1 tubule. (b) Overlaid surface view of the density maps of the largest diameter class (magenta) and smallest diameter class (blue). Note the color dominance of the small diameter class along the inside of the tubule, illustrating its smaller diameter. (c) Zoom-in view of a region of the cryoEM density map of the NS1 tubule at 4.0A. Individual monomers are colored differently to distinguish borders of interacting monomers. Monomer-monomer interfaces A and B are indicated.
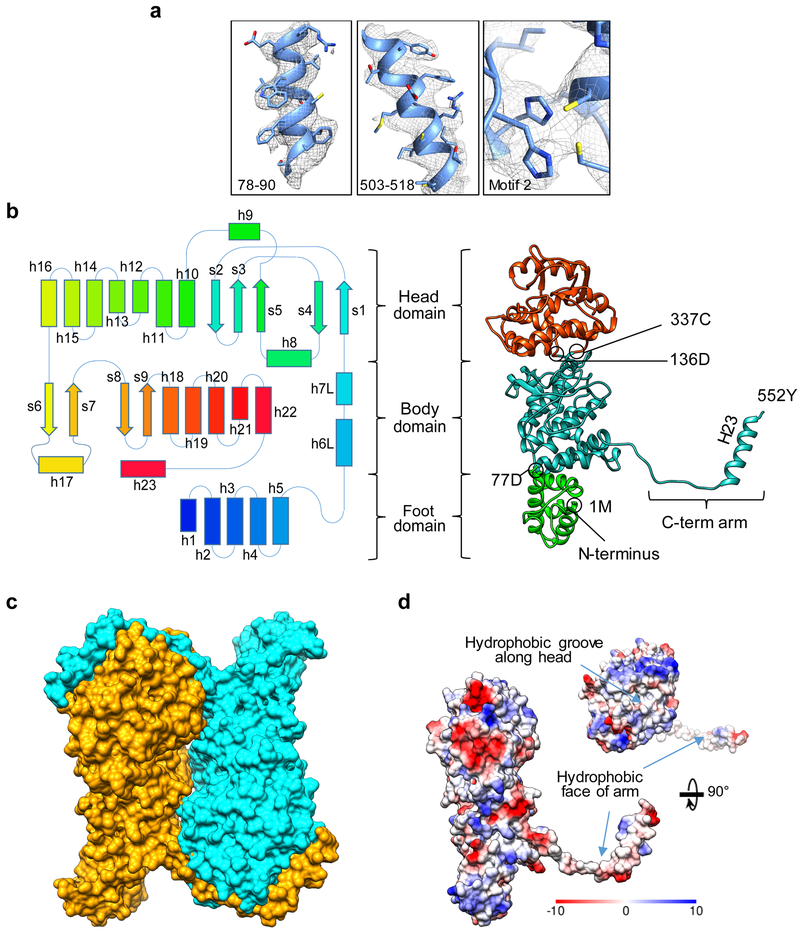
Figure 2: Structure of the NS1 monomer.
(a) Superposition of the cryoEM densities (gray mesh) and their corresponding atomic models (ribbon and sticks) for three selected regions of NS1 illustrating the quality of the cryoEM densities that supports atomic modeling based on amino acid side chains. (b) Secondary structure schematic and domain architecture (left) mapped to the atomic model (shown as ribbon diagram to the right) of an NS1 Monomer. (c) Space-filling surface rendering of an NS1 dimeric building subunit of both tubular forms. (d) Coulombic surface rendering of the NS1 monomer showing the C-terminal arm handshake. Note the hydrophobic groove in the head domain and the hydrophobic inner surface of the C-terminal arm.
Structure reveals that the 2-start helical tubule is formed by pseudo-planar NS1 homodimers arranged into a thin-walled (~57 Å) tubule, containing 41.16 monomers/turn. Its architecture is stabilized laterally by packing interactions between neighboring NS1 subunits of the same layer, and vertically by compact junctions between six monomers from two different layers (Fig. 1c). This helical arrangement differs from that reported previously9. Remarkably, the 1-start/2-start helical change in the tubule arrangement does not alter the monomeric structure in a noticeable manner, reminiscent of the formation of a carbon nanotube from graphene11. NS1 monomers interact with its neighbors through two interfaces, A and B (Fig. 1c). The dimers are packed together in the 1-start in an upright position with respect to the tubule axis (the angle between the dimer interface and the tubule axis is 18.8° and 20.7° in the 1-start and 2-start tubules, respectively). There are no major changes in subunit-subunit interactions between the two helical forms, and only a subtle shift in the arrangement of each subunit with its neighbors. These similarities and differences indicate an assembly mechanism of an NS1 tubule from a flat and flexible sheet, a process that allows different helical configuration to form.
Structure of the NS1 monomer
NS1 has no known structural homologs, and its repeating arrangement with entwined extensions in the tubule created difficulties in de novo modeling. Some prominent structural features, such as aromatic side chains (Fig. 2a), and the visible C-terminus provided key landmarks for model building. Subsequently, integration of structural map with sequence-based prediction and available functional data allowed us to complete a provisional model consisting of two of the three globular domains of the NS1 monomer. In retrospect, resolving key junctions at domain boundaries was challenging due to confusion caused by the previously unknown presence of metal ion coordination motifs. Such motifs created density branches obscuring chain tracing. Once these two globular domains were assigned, the remaining globular domain and the C-terminal arm became readily assignable to the correct monomer, allowing de novo modeling and refinement of the first NS1 atomic model (Fig. 2b).
Each NS1 monomer is organized into three domains (Fig. 2b): foot, body and head, with the N-terminus located in the foot domain and the C-terminus extending far from the body domain through an “arm”. Monomers are arranged in the tubule in an alternating head-to-foot orientation (Fig. 2c). The head domain consists of nine α helices and two β sheets. One of the β sheets contains anti-parallel strands and the other, both anti-parallel and parallel strands. The body domain lies between the head domain and the foot domain. It is comprised of two β sheets and six α helices. The foot domain (residues 1–77) is composed of five α helices, with two linker helices h6L and h7L traversing the body domain to reach the head domain (residues 136–337).
The C-terminal arm is necessary for tubule formation but not for virus replication
The NS1 C-terminal arm contains a 15-residue long alpha helix (h23, residues 535–550), and connects to the globular body domain through helix 22 upstream (Fig. 2b). The hydrophobic arm extends from the body and occupies a hydrophobic groove on the neighboring monomer while its terminal helix displays a hydrophobic face to grasp the head domain of the neighboring monomer (Fig. 2c & 2d). Each monomer reaches out to join the neighboring monomer and in turn, is inter-linked by its partner (Fig. 2c). Furthermore, the hydrophobic β-strand section (residues 522–526) of the C-terminal arm augments a β sheet in the head domain of the neighboring monomer formed by strands s2, s3, and s5 (Fig. 3a).
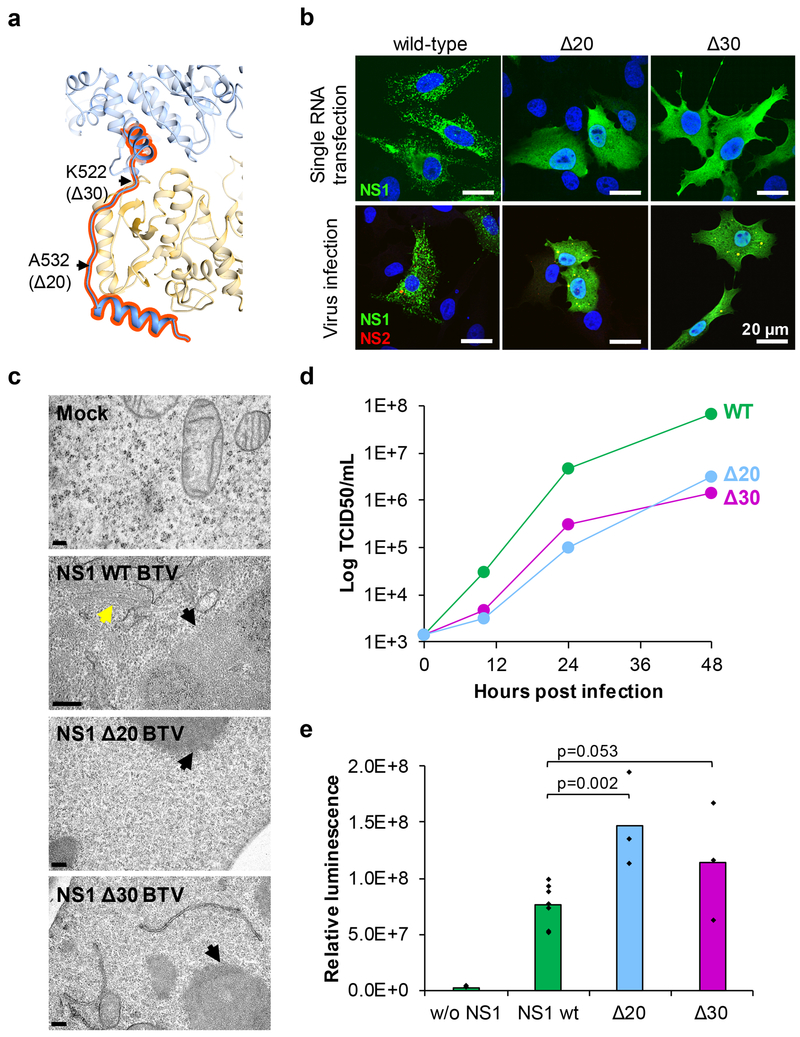
Figure 3: C-terminal arm deletion mutations demonstrate that the non-tubular form of NS1 is the functional form.
(a) Ribbon diagram inter-NS1 interface showing the locations of the two C-terminal arm deletion mutants. The β sheet augmentation and handshake grip region of the C-terminal arm are highlighted in red. Residues for deletion mutants are labeled (K522 and A532). (b) Immunofluorescence analysis showing the intracellular localization of wild-type or mutated NS1 (Δ20 and Δ30) in cells transfected with NS1 capped mRNA (top panels) or infected by mutant viruses (bottom panels). NS1, green (rabbit anti-NS1 primary antibody and anti-rabbit Alexa 488 coupled secondary antibody); NS2, red [component of the viral inclusion bodies (VIBs); mouse anti-NS2 primary antibody and anti-mouse Alexa 546 coupled secondary]; Nuclei, blue (Hoechst staining). Scale bars, 20 μm. Experiments were repeated twice independently with similar results. (c) Transmission electron microscopy analysis of sections of cells infected with wild-type and mutant viruses as indicated. VIBs are indicated (black arrows). Tubules (yellow arrow) are only present in the cytoplasm of cells infected by the wild-type virus. Scale bars, 200 nm. Sectioning data is the result of one experiment. (d) Growth curves of viruses. Virus titers were determined in triplicates. (e) Quantification of Renilla luciferase expression by wild-type or mutant NS1. Luciferase activity was detected 24h after cells were co-transfected with S10-Rluc and wild-type or mutated NS1 capped mRNA. Bars represent the averages of at least three independent experiments (Wild-type n=7, mutants n=3). Distribution of the data and respective p-values are indicated (one-tailed t test: two-sample assuming equal variance, confidence level of 95%).
Previous biochemical studies12 have shown that the C-terminal 10 amino acids are essential for tubule formation, suggesting that the C-terminal helix is involved in tubule formation. To investigate this, two C-terminal deletion mutants, Δ20 (Δ532–552) and Δ30 (Δ522–552) were generated using NS1-encoding gene S5 (Fig. 3a) and their effects on virus replication were assessed using reverse genetics13. Both mutant viruses were recovered successfully (Fig. 3b), indicating that virus replication was not significantly hampered. However, we did not observe the characteristic cytoplasmic distinct granular distribution of NS1 (left panels in Fig. 3b) exhibited in wild-type transfected or virus-infected cells, but rather diffuse patterns of NS1 present throughout the cytoplasm (middle and right panels in Fig. 3b). However, there was no obvious difference in the plaque phenotype between mutant viruses and the wild-type virus (Fig. S4), although no tubules were observed by electron microscopy of infected cells (Fig. 3c). Nonetheless, the growth rate of the mutant viruses decreased slightly compared to the wild-type (Fig. 3d). Using a reporter RNA assay4, we tested both Δ20 and Δ30 for their ability to specifically enhance viral protein expression as described in Methods. In each case, expression of the reporter was detected (Fig. 3e), indicating that the NS1 function was not perturbed by the C-terminal deletion. The recovery of these mutant viruses demonstrates that the C-terminal arm and its β-sheet augmentation role are required for tubule formation, but not for the function of the NS1 in protein expression, demonstrating for the first time that the non-tubular form of NS1 is functional. The characteristic shape of this C-terminal arm linking another monomer suggests that it facilitates dimer formation.
NS1 contains two zinc-finger-like motifs that are important for virus replication and tubule formation
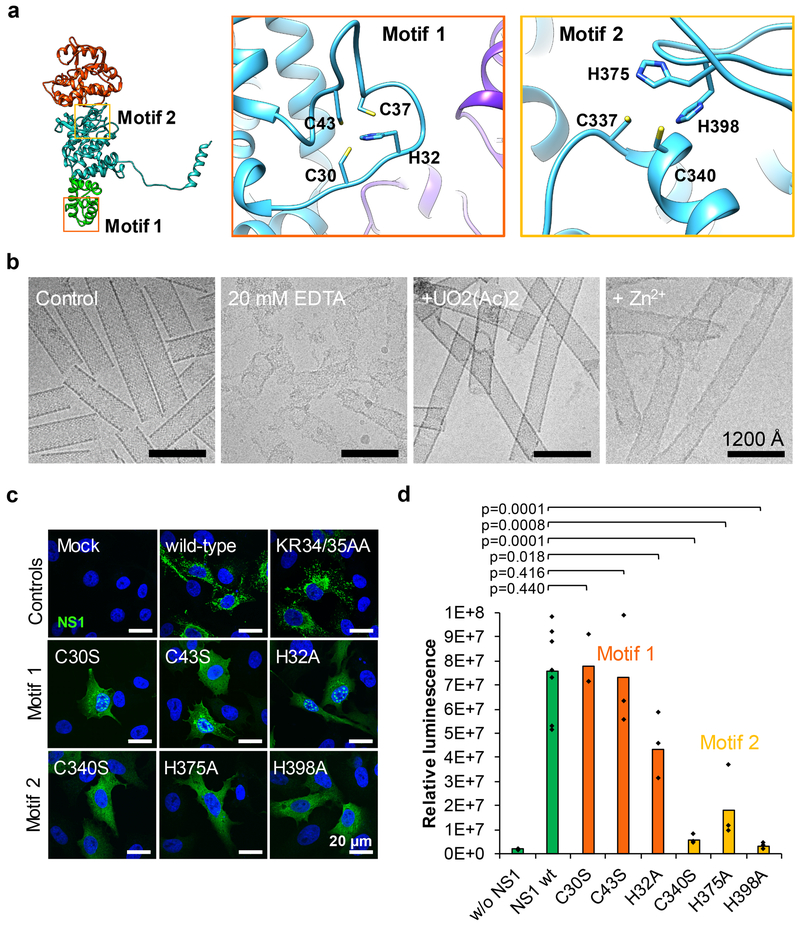
Each NS1 monomer contains two zinc-finger-like motifs, Motif 1 and Motif 2 (Fig. 4a). Both motifs have tetrahedral arrangement of side-chain rotamers and a strong density at the center of the tetrahedron, characteristic of metal coordination in typical zinc-finger motifs. The putative metal ion coordination site of Motif 1 is formed by a tetrahedral arrangement of C30, H32, C37, and C43 (Fig. 4a, orange panel). In the tubular form, pairs of foot domains, head domains, and C-terminal arms cage Motif 1 in tubular NS1, rendering it inaccessible from both outside and inside the tubule. In contrast, Motif 1 in both NS1 monomer (Fig. 4a) and dimer (Fig. 2c) forms are readily accessible. The tetrahedral metal ion coordination site of Motif 2 is located at the junction between the head and body domains and is formed by C337, C340, H375, and H398 (Fig. 4a, yellow panel). Its secondary structure arrangement is reminiscent of, but distinct from, typical Cys2His2 zinc fingers, which exhibit a αββ architecture. H375 and H398 reside on separate strands of a three-stranded β sheet, and are connected by a loop and strand. C337 and C340 are located on h16 at the base of the head domain.
Figure 4: NS1 contains two putative zinc-finger motifs, which are important for virus replication and tubule formation.
(a) Ribbon diagram of an NS1 monomer showing the locations (left) and details (insets) of putative metal ion binding Motifs 1 and 2. The four residues implicated in the tetrahedral coordination in each motif are labeled in the insets. (b) CryoEM micrographs of well-structured tubules (control), of disrupted tubules in the presence of 20 mM EDTA, and of tubules recovered upon addition of divalent uranyl and zinc ions. Experiment was repeated twice independently with similar results. (c) Immunofluorescence analysis showing the intracellular localization of wild-type or mutated NS1 (Motif 1: C30S, C43S, H32A; Motif 2: C340S, H375A, H398A) in cells transfected with the respective capped mRNA. The KR34-35AA mutant was used as a control. NS1, green (rabbit anti-NS1 primary antibody and anti-rabbit Alexa 488 coupled secondary antibody); Nuclei, blue (Hoechst staining). Scale bars, 20 μm. Experiments were repeated twice independently with similar results. (d) Quantification of Renilla luciferase expression by wild-type NS1 or NS1 harboring putative zinc-finger motif mutations. Luciferase activity was detected 24h after cells were cotransfected with S10-Rluc and wild-type or mutated NS1 capped mRNA. Bars represent the averages of at least three independent experiments (Wild-type n=7, mutants n=3). Distribution of the data and respective p-values are indicated (one-tailed t test: two-sample assuming equal variance, confidence level of 95%).
To verify that these motifs are divalent metal ion coordination motifs, we treated tubules with chelating agent and examined structural changes. The integrity of tubules was compromised by such treatment, leading to fluffy aggregates, which can be restored by divalent cations, including Zn2+ (Fig. 4b). This observation indicates that the presence of divalent metal ions may be important for the maintenance of tubular NS1 and that the conversion between the tubular and non-tubular forms is reversible by removal and addition of metal cations.
To determine the importance of these putative zinc-binding motifs, substitution mutations (C30S, C43S, H32A in Motif 1 and C340S, H375A and H398A in Motif 2) were generated by site-directed mutagenesis in viral genome. Each mutation in Motif 1 and Motif 2 failed to recover viruses by reverse genetics, although a virus was recovered successfully when nearby residues were mutated (KR34–35AA) (Fig. S4). Expression of NS1 in each case was visualized by immunofluorescence, although their cytoplasmic distribution was diffused (middle and bottom rows in Fig. 4c). In contrast, the wild-type NS1 and the positive control KR34–35AA NS1 mutant showed the same characteristic cytoplasmic distribution (top row, Fig. 4c). Given that zinc fingers are often involved in nucleic acid binding, it is possible that either of the metal ion coordination motifs is important for structural stability and/or related to the ability of NS1 to selectively upregulate viral mRNA in host cells4. However, mutations in Motif 1 had no significant effect on the upregulation of the reporter expression, except the H32A mutant, which showed only a slight decrease in the transfected cells (Fig. 4d, orange bars). In contrast, all mutations in Motif 2 had drastic effects (up to 21 fold) on the luciferase expression (Fig. 4d, yellow bars), indicating Motif 2 is particularly important to upregulate viral gene translation, although both metal ion coordination motifs are necessary to maintain the characteristic tubular structure.
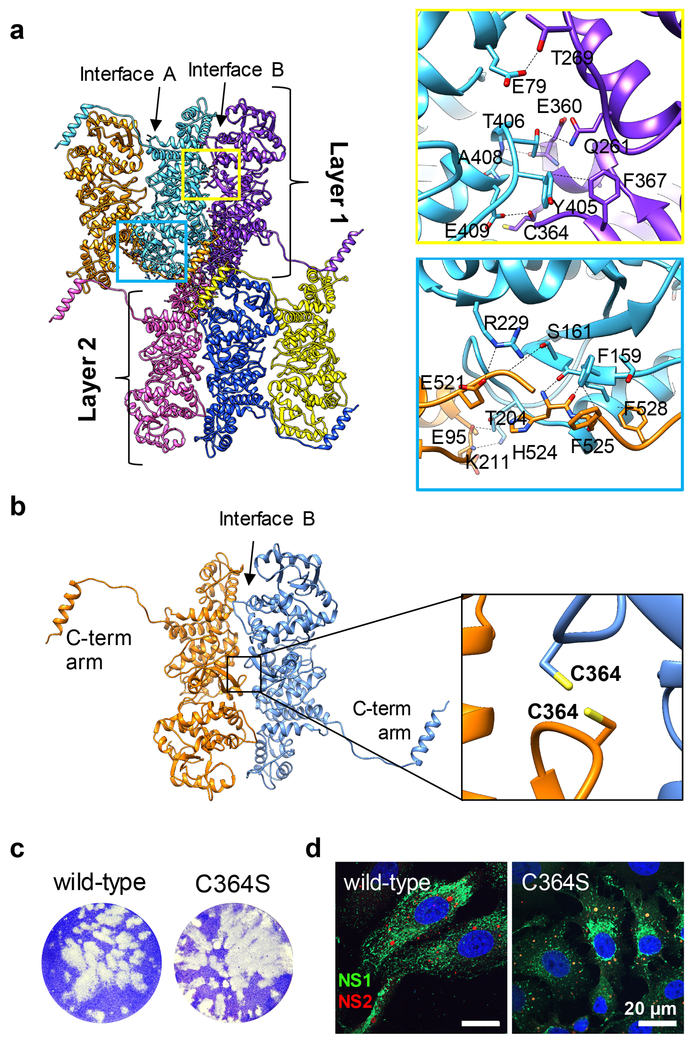
Intra-layer monomer-monomer interactions
Two large binding surfaces spanning the length of the NS1 protein are presented by each monomer to its two neighboring monomers within the same layer. These interfaces (A and B) include a mixture of hydrophobic and charged residues (Fig. 1c and Fig. 5a), dictating extensive interactions between monomers of the same layer. The interface B revealed many interactions including a possible disulfide bond formation between C364 of one monomer and C364 of the other monomer in the same layer around a sideway two-fold symmetry axis (Fig. 5b). Previous studies using recombinant NS1 protein showed that a C364S mutant formed tubules similar to wild-type NS114. To investigate in the context of virus infection, we created the same mutation in the viral genome and successfully recovered C364S mutant virus with a similar plaque phenotype to the wild-type (Fig. 5c). Cells infected with plaque-purified wild-type or C364S mutant viruses showed efficient viral replication although the intracellular localization of NS1 C364S was slightly affected (Fig. 5d). Taken together, these results indicate that such a disulfide bond is unlikely to be critical to tubule formation and suggest that interface B is complex and will need further studies.
Figure 5: Inter- and intra-layer interactions in NS1 tubules.
(a) Interlayer six-monomer junction in NS1 tubule. Blue inset, atomic models showing intra-layer beta-sheet augmentation involving the head domain of one monomer and the C-terminal arm of the other monomer. Yellow inset, atomic models of sample intra-layer packing between the head domain of one monomer and the foot domain of another monomer. Monomer-monomer interfaces A and B are indicated. (b) Putative disulfide bond between monomers of the same layer. Black inset: atomic models of C364 sidechain position and orientation. (c) Recovery of C364S mutant virus by reverse genetics as compared to the wild-type virus. Plaques (white) indicative of recovered viruses, were visualized by crystal violet staining. Similar plaque phenotypes were observed for the wild-type and mutant viruses. (d) Intracellular localization of wild-type or C364S mutant NS1. Immunofluorescence analysis showed similar VIBs (indicative of similar viral recovery) but partially affected intracellular NS1 localization in cells infected with the mutant virus as compared to those infected with the wild-type virus. NS1, green (rabbit anti-NS1 primary antibody and anti-rabbit Alexa 488 coupled secondary antibody); NS2, red [component of the viral inclusion bodies (VIBs), mouse anti-NS2 primary antibody and anti-mouse Alexa 546 coupled secondary antibody]; Nuclei, blue (Hoechst staining). Scale bars, 20 μm. (c, d) Experiments were repeated twice independently with similar results.
Discussion
In this study, we obtained the first atomic model of BTV NS1, derived from a near-atomic resolution cryoEM helical reconstruction of NS1 tubules. The observation of multiple, dynamic classes of tubules indicate that NS1 monomers are able to assemble with some degree of flexibility, without affecting the monomeric structure. This ability to form tubules of variable helical forms and diameter suggests that tubule formation is a robust process capable of initiating in various configurations. The helical reconstruction shows that the immunogenic carboxyl terminus is located along the tubule surface, consistent with several previous studies, which indicated to this localization12,15. The location of the carboxyl terminus rationalizes the ability of NS1 tubules to serve as effective immunogen delivery vehicles, capable of carrying large peptides without disrupting the tubular structure15–17. The structure-based mutagenesis studies showed that deleting NS1 C-terminal arm abrogated tubule formation but still could recover infectious virus and retained ability to regulate viral protein expression as confirmed by a reporter assay. Thus, the non-tubular form of NS1 is sufficient for virus replication and for upregulating viral mRNA translation. While the tubular form is not required for upregulating viral RNA translation, tubule formation during BTV infection had been reported to reduce cytopathic effects, suggesting that NS1 tubules may play a role in cellular pathogenesis18,19.
Our NS1 structure shares no recognizable similarity with any published structures. During rotavirus infection, the non-structural viral protein NSP3A acts like cellular poly(A) binding protein 1 (PABP1) and binds to the 3′ end of viral mRNA2. Motifs implicated in dimerization, RNA binding and interaction with EIF4G1 have been mapped in rotavirus NSP33, and although both N-terminal and C-terminal domain structures of NSP3 were described, neither significant sequence identity nor structural similarity between BTV NS1 and rotavirus NSP3 can be recognized7,8. A human cellular protein conserved in mammals, termed rotavirus “X”-associated non-structural protein (RoXaN), has been shown to form a ternary complex with eIF4G and rotavirus NSP35, thereby promoting interaction between cytoplasmic poly(A) binding protein and viral RNA6. Surprisingly, RoXaN bears some sequence similarity to BTV NS1 at the solvent-exposed regions except for the C-terminal arm, with 94 identical and 177 similar amino acid residues (Fig. S2). Given that NS1 is known to selectively upregulate viral protein synthesis by binding to the 3’ end of viral mRNA4, we speculate that NS1 may mimic some function of RoXaN, plausibly promoting interaction between viral mRNA and eIF4G or other components of host translation machinery in order to upregulate viral protein synthesis.
Our atomic structure of BTV NS1 reveals two zinc-finger-like motifs with tetrahedral arrangements, Motif 1 and Motif 2, and our functional studies revealed that tubule structure was significantly affected by chelating agent treatment. Remarkably, RoXaN possesses five similar motifs (C3H1 and C2H2 types). The first of these five motifs is localized to a region with 100% identity with BTV NS1’s Motif 2 (337-CQLCY-341, Fig. S2b), which is necessary for NS1 to fulfill its upregulation function of viral mRNA translation. By contrast, the region around Motif 1 of BTV NS1 aligns with RoXaN only in a limited fashion Fig. S2b). Zinc finger motifs are known to stabilize protein structure, or to bind nucleic acids, or to mediate protein-protein interactions20,21. Our data show that while Motif 2 plays a major role for the upregulation of viral protein translation, the integrity of both motifs is likely essential for maintaining the stability of NS1 structure. In addition, typical granulated patterns observed in the cells transfected with wild-type NS1 were not present in the mutant NS1 transfected cells, consistent with our conclusion that these motifs are likely to be important for the tubule stability. However, how NS1 zinc-finger motifs up-regulate BTV replication awaits further investigation. Nevertheless, we do not rule out possibilities that these motifs bind alternatively some other, unidentified metal ions22. Selection pressure has forced organisms to evolve complex regulation processes for gene expression regulation, through multiple levels of control and involving many molecules of nucleic acids and proteins. Facing the scarcity of resources, a simple organism such as a virus has to employ different strategies to regulate its gene expression, including using multiple oligomeric states of a single protein. Considering the fact that the tubules, purified from BTV-infected cells, assemble in variable helical forms that can be converted to non-helical forms by the level of zinc or other divalent cations, we hypothesize that BTV might employ a negative feedback mechanism to regulate viral protein translation by sequestering the functional non-tubular form of NS1 into tubular assemblies. This process may involve regulation of local concentration of metal ions. As BTV is a model virus system for many dsRNA viruses, these findings offer a rare glimpse into the regulatory aspect of viral life cycle23, while simultaneously providing a trove of information on both tubular and non-tubular forms of NS1 and the relationship between the two forms. It opens the door to new possibilities in structure-function relationship studies to allow further understanding of NS1-host cell machinery interaction.
Material and methods
Cells and viruses
BSR cells (BHK-21 subclone) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% (v/v) foetal bovine serum (FBS) at 35°C in 5% CO2.
Wild-type and mutant BTV viruses were recovered by reverse genetics as previously described13 (see below). Each recovered virus was plaque-purified, amplified in BSR cells and harvested at 100% cytopathic effect (CPE) between 2–3 days. Viruses were titrated either using plaque assay or TCID50 assay.
Tubule purification
BSR cells were infected with wild-type or mutant BTV1 at MOI (Multiplicity of Infection) 0.5 during 1h. Viral inoculum was subsequently removed and cells were incubated in 1% FBS culture medium for 44h until 100% CPE. Supernatant and cells were harvested and clarified 10min at 4500 rpm. Cell pellets were resuspended in prechilled lysis buffer (100mM Tris HCl pH 7.5, 50mM NaCl, 10mM EDTA, 0.1% NP-40 [Sigma]), incubated at 4ºC for 10min and spun down at 4000 rpm 5min at 4ºC. Supernatants were kept on ice and lysis was repeated once. Supernatants were pooled and loaded on a 40% w/v sucrose cushion in 20mM Tris HCl pH 7.5, 150mM NaCl buffer. Ultracentrifugation was carried out at 28,000 rpm at 4ºC for 2h. Pellet was resuspended overnight in 20mM Tris HCl pH 7.5, 150mM NaCl buffer prior to vitrification for cryoEM sample preparation.
CryoEM
Optimization for sample distribution and ice thickness was performed in an FEI TF20 cryo electron microscope equipped with a TVIPS 16 MegaPixel CCD camera, first by negative stain and then by cryoEM. High-resolution cryoEM images were recorded in a Titan Krios instrument over a 10 years of period in an effort to improve the resolution of the structure. Two different types of cryoEM micrographs were recorded and processed in this effort. Prior to the advent of direct electron detectors, we recorded cryoEM images on photographic films (film dataset). However, after exhaustive efforts and 2,087 films, we were unable to improve our 3D structure to be better than 4.5 Å resolution. For this reason, when electron counting became available to us, we imaged the same tubules again in a Gatan K2 Summit direct electron camera attached to an energy filter. Direct electron detector dataset of cryoEM images were collected as movies in an FEI Titan Krios microscope (operated at 300kV) equipped with a Gatan imaging filter (GIF) (the slit was not inserted) and K2 Summit direct electron camera in counting mode using Leginon software package24 for automation. Target defocus value is set to 2.0 μm under focus. Each movie contains 50 frames with 5 frames per second with a total accumulated dosage of 60 electrons per Å2. The dose rate is measured at 6 electrons per Å2 per second in the Digital Micrograph software package, which is calibrated to 7.5 electrons per Å2 per second compared to our initial efforts with films25,26. A total of 5,006 movies were collected over two sessions.
Image processing for the film data
Micrographs recorded on Kodak SO163 photographic films were digitized and screened to select tubules that are non-overlapping, intact and free of ice-contamination. Tubule particles were manually selected with EMAN27 helixboxer with a box width of 640 and was segmented according to a 10% overlapping scheme. A total of 218,238 segments were selected from images from all sessions. The contrast transfer function (CTF) parameters of these movies were determined by CTFFIND328. The determined defocus range was 0.6–3.1 μm.
We carried out reference free 2D classifications (tier I) of these particles by EMAN refine2d.py. The resulting class-averages show different tube diameters. We further classified these classes manually by their diameters (4 different diameters). For each meta-class (tier II), all class-average images were transformed into Fourier space so that their amplitude images (layer line images) were translated into real space images. These images were classified again in 2D (tier III, 10 classes). For every tier II class, we noticed that there are three modes (tier IV) of peri-meridian reflections in the 10 (tier III) class-averages. These four tiered classification suggested that there are 12 (4×3) different helical forms in the population of particles.
We indexed every tier IV class-average as previously described by David DeRosier (http://www.biomachina.org/courses/structures/download/derosier_handout_02.pdf) and Ref 29. Basically the three modes of peri-meridian reflections put (0,1) at n = 1, 2 and 3 respectively (one, two and three-start helices). Surprisingly, for all helices, the pitches are roughly the same, being around 88 ~ 89 Å; however, their l numbers are different. The selection rule for all three helical form of class 2 diameter helices (tier II), as determined from (0,1) and (1,0) reflections are (l=−5n+101m; n=20, l=1 and n=−1, l=5), (l=−7n+144m; n=20, l=4 and n=2, l=14) and (l=−11n+232m; n=20, l=12 and n=−3, l=33) (Supplementary Fig. S3). Other helical forms can be deduced in a similar way.
We sorted the particles according to their tier IV class identities and calculated a volume for each of them by EMAN based IHRSR25,30. Further image processing steps were performed with Relion v1.2 with IHRSR31. The selected particles for tier II class 2 – tier IV class 1 (20.2 dimers per turn, tier IV grand rank #4), tier II class 2 – tier IV class 2 (20.57 dimers per turn, tier IV grand rank #5), tier II class 3 – tier IV class 1 (21.2 dimers per turn, tier IV grand rank #7) and tier II class 2 – tier IV class 2 (21.57 dimers per turn, tier IV grand rank #8) were refined with Relion v1.2 respectively. The final resolutions for resulting maps are in the 4.5 – 5.0 Å range, marginally unsuitable for atomic modeling.
Image processing for the direct electron detector data
Frames within each movie were aligned to correct for drift as previously described32, except that an iterative alignment scheme was used as previously described elsewhere 33. Aligned frames were averaged to generate two average micrographs for different purposes: one by averaging 1st to 50th frames for particle selection and determination of contrast transfer function (CTF) parameters, and the other by averaging 3rd to 20th frames for structure refinement. CTF parameters were determined by CTFFIND3f28 and those with very large and very small underfocus values were discarded. The determined defocus ranges for included data were 1.2 ~ 4 μm for session 1 and 1.5 ~ 3.5 μm for session 2.
Tubule particles were manually selected the same way as with film dataset except for a 720 box width and was segmented according to a 10% overlapping scheme; a total of 87,190 segments were selected for images from both sessions. We cleaned up the particles by using Relion v1.4 Class2D.
For each session, all particles were subjected to a Class3D run with 12 classes for the separation of the 12 helical forms. For each helical form, particles from the two sessions were combined. We further analyzed the helical forms of 20.2 dimers per turn (grand rank #4) and 20.57 dimers per turn (grand rank #5), respectively.
For each of the two selected helical forms, a Class3D run with five classes was done to separate particles at different stages of thermal breathing of the tube. Only the class with the most particles—incidentally the class with the medium diameter—was included for further processing, respectively.
We initially used Refine3D in Relion v2.0 to refine the structures. However, given the SNR based regularization intrinsic to the Relion algorithm, a working resolution control was not satisfactory, since there are too many helically related copies and SNR compensation that worked for previous studies with less helically related copies31 did not work properly. We therefore used Class3D with one class to refine the structures and manually adjusted T factor (--tau_fudge) and healpix order so that the refinement could progress.
We then first averaged the two monomers in the dimers from the 1-start helical form with Chimera34. The monomers were automatically averaged in the 2-start helical form since dihedral (D2) symmetry were imposed during refinement. We then averaged the dimers across two helical forms with Chimera34.
Atomic model building
The backbone of the NS1 monomer was traced using Coot35. PHYRE236 prediction server was used to partially guide backbone tracing. The backbone coordinates were then changed to peptide and subsequently mutated to the correct sequence. The initial N-terminus assignment was corrected to account for a two-fold symmetry axis. The subsequent model went through manual regularization using Coot37, before undergoing real-space-refinement through PHENIX38. The output was analyzed using the Molprobity39 integration in Coot and had its clashscore and geometry improved manually. The structure was then subjected to geometry_minimization in PHENIX and then fed to real-space-refinement with simulated annealing, using MolProbity functions built into PHENIX to track model quality and agreement with the density map throughout refinement. The final model was checked for fitting errors manually in Coot and UCSF Chimera34. All structural figures were generated using UCSF Chimera.
Plasmids
Plasmids used in this study were pCAGGS BTV1 protein expression plasmids (pCAG-VP1, pCAG-VP3, pCAG-VP4, pCAG-VP6, pCAG-NS2) and T7 plasmids for BTV transcripts as previously described13.
Site-directed mutagenesis
Site-directed mutagenesis of BTV1 NS1 was performed using the T7 segment 5 (encoding NS1) template and the following mutagenic primers (5’−3’):
C30S (CCACAATGGACTAGCAGTCATCTAAAAAGG),
C43S (GGAATTGTTTATTCAATGGGATGAGTGTTAAACAAAATTTTGAGAGAGC),
H32A (CAATGGACTTGCAGTGCTCTAAAAAGGAATTG),
KR34/35AA (GGACTTGCAGTCATCTAGCAGCGAATTGTTTATTCAATGG),
C340S (GCATACATGTCAGCTGAGCTACTTGAAACACTC),
C364S (CATCAGAACTAACTGGGTCTTCGCCATTCAAGACGG),
H375A (GTGAAGATTGAGGAAGCTGTGGGAAATGATTCG),
H398A (GGCAGGATCGGAGATGCTTATTATACTACAAATTG)’
Delta20 (GCTGGGTTCGCGGCACCTGCGTAGTTACTGACTTCTGTT),
Delta30 (CCCTATGCTATGCAGAAAAGTAGTTACTGACTTCTGTTTTCTG).
Mutagenic bases are underlined. Obtained mutants were subsequently sequenced using an internal NS1 primer in order to confirm the presence of the desired mutation(s).
Synthesis of NS1 capped mRNAs
Synthetic single-stranded RNAs were prepared by runoff in vitro transcription from T7 PCR products using T7 RNA polymerase. Transcripts were prepared with anti-reverse cap analogue (ARCA) using the mMESSAGE mMACHINE T7 Ultra kit (Ambion) as previously described13. Transcripts were analysed by electrophoresis in 1% agarose in morpholinepropanesulfonic acid (MOPS) in the presence of formaldehyde.
Transfection of BSR cells
Seventy percent confluent BSR monolayers in 12 well plates were transfected with 500 ng of mRNA transcripts using Endofectin (GeneCopoeia), according to the manufacturer’s instructions. Cells were subsequently incubated at 35°C in 5% CO2 8 to 24h after transfection.
Reporter RNA transcripts preparation and quantification of reporter expression
Reporter RNAs were prepared and the quantification of reporter expression was performed as previously described4 with slight modifications. Briefly, the pS10-Rluc plasmid clone containing the 5′ 152nt and 3′ 149nt of BTV-1 segment 10 fused with the Renilla luciferase gene was digested with restriction enzymes, and synthetic capped single-stranded RNAs were prepared by run-off in vitro transcription using the mMESSAGE mMACHINE T7 Ultra kit (Ambion). BSR 96-well monolayers were transfected with 150 ng of capped S5 RNA wild-type or mutant and 150 ng of capped S10-Rluc reporter RNA. Renilla luciferase expression in the transfected cell lysates was quantified in triplicates 24 h after transfection with a Turner Biosystems Glomax luminometer with Promega’s dual luciferase reporter assay system according to the manufacturer’s instructions.
Recovery of viruses by reverse genetics
Reverse genetics was performed as previously described13. Briefly, at day 1, 70–80% confluent BSR monolayers were transfected with pCAG-VP1, pCAG-VP3, pCAG-VP4, pCAG-VP6 and pCAG-NS2 (120 ng each) using Endofectin (GeneCopoeia), according to the manufacturer’s instructions, and incubated at 35°C in 5% CO2 overnight. At day 2, the cells were transfected with each BTV1 exact copies RNA transcripts (S5 wild-type or mutated) using Endofectin (GeneCopoeia), overlaid with 1% agarose and incubated 3 days at 35°C in 5% CO2. Visible plaques were picked up and resuspended in 1% FBS containing medium, and/or cells were subsequently fixed with 10% formaldehyde and stained with crystal violet.
Immunofluorescence assays
BSR cells were grown on coverslips and either transfected with capped mRNA or infected with wild-type or mutant viruses. Eight to ten hours post transfection or infection cells were fixed with 4% paraformaldehyde (Sigma) solution, permeabilized with 0.5 % Triton X100 (Sigma), blocked with 1% BSA (Bovine Serum Albumin, Sigma), and subsequently stained using rabbit anti-NS1 and/or mouse anti-NS2 primary antibodies, and anti-rabbit Alexa 488 or anti-mouse Alexa 546 coupled secondary antibodies (Thermo Fisher Scientific). Nuclei were stained using Hoechst 33342 (Thermo Fisher Scientific). Images were acquired using an x100 oil objective and a Zeiss Axiovert LSM510 confocal microscope supplied with the LSM510 software.
Cell sectioning analysis
BSR cells were infected at MOI 5 and were processed for cell sectioning at 16 h post infection. Briefly, monolayers were fixed in 2% paraformaldehyde-1.5% glutaraldehyde-0.1M sodium cacodylate buffer (pH 7.3) and postfixed in 1% osmium tetroxide-1.5% potassium ferrocyanide-0.2M sodium cacodylate buffer. Cells were dehydrated in increasing concentrations of ethanol and embedded in epoxy resin (TAAB Laboratories Equipment Ltd., UK). Ultrathin sections were stained with lead citrate.
Supplementary Material
Acknowledgements.
We thank Mark Turmaine for his advice and support for imaging at the UCL EM facility and Cristina Celma (LSHTM) for advising in BTV reverse genetics method. This project is supported partly by grants from the National Institutes of Health (AI094386 to ZHZ) and The Wellcome Trust, UK (100218, Investigator Award to PR). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and grants from NIH (1S10OD018111 and 1U24GM116792) and NSF (DBI-1338135 and DMR-1548924). This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562 (Comet cluster at the San Diego Supercomputing Center through allocation MCB140140).
Footnotes
Data availability: The atomic models and cryoEM density map that support the findings of this study have been deposited in PDB and EMDB with accession numbers 6N9Y and EMD-0383, respectively.
References:
- 1.Walsh D & Mohr I Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9, 860–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Piron M, Vende P, Cohen J & Poncet D Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J 17(1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piron M, Delaunay T, Grosclaude J & Poncet D Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J Virol 73, 5411–21 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce M, Celma CP & Roy P Bluetongue virus non-structural protein 1 is a positive regulator of viral protein synthesis. Virology Journal 9, 178 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitour D, Lindenbaum P, Vende P, Becker MM & Poncet D RoXaN, a novel cellular protein containing TPR, LD, and zinc finger motifs, forms a ternary complex with eukaryotic initiation factor 4G and rotavirus NSP3. J Virol 78, 3851–62 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harb M et al. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82, 11283–93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deo RC, Groft CM, Rajashankar KR & Burley SK Recognition of the rotavirus mRNA 3’ consensus by an asymmetric NSP3 homodimer. Cell 108, 71–81 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Groft CM & Burley SK Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol Cell 9, 1273–83 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Hewat EA, Booth TF, Wade RH & Roy P 3-D reconstruction of bluetongue virus tubules using cryoelectron microscopy. J Struct Biol 108(1992). [DOI] [PubMed] [Google Scholar]
- 10.Urakawa T & Roy P Bluetongue virus tubules made in insect cells by recombinant baculoviruses: expression of the NS1 gene of bluetongue virus serotype 10. J Virol 62(1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odom TW, Huang JL & Lieber CM Single-walled carbon nanotubes: from fundamental studies to new device concepts. Ann N Y Acad Sci 960, 203–15 (2002). [PubMed] [Google Scholar]
- 12.Monastyrskaya K, Gould EA & Roy P Characterization and modification of the carboxy-terminal sequences of bluetongue virus type 10 NS1 protein in relation to tubule formation and location of an antigenic epitope in the vicinity of the carboxy terminus of the protein. J Virol 69, 2831–41 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyce M, Celma CC & Roy P Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J Virol 82, 8339–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monastyrskaya K, Booth T, Nel L & Roy P Mutation of Either of 2 Cysteine Residues or Deletion of the Amino or Carboxy-Terminus of Nonstructural Protein Ns1 of Bluetongue Virus Abrogates Virus-Specified Tubule Formation in Insect Cells. J Virol 68, 2169–2178 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikhailov M, Monastyrskaya K, Bakker T & Roy P A new form of particulate single and multiple immunogen delivery system based on recombinant bluetongue virus-derived tubules. Virology 217, 323–31 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Ghosh MK, Borca MV & Roy P Virus-derived tubular structure displaying foreign sequences on the surface elicit CD4+ Th cell and protective humoral responses. Virology 302, 383–92 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Murphy A & Roy P Manipulation of the bluetongue virus tubules for immunogen delivery. Future Microbiol 3, 351–9 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Owens RJ, Limn C & Roy P Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. Journal of Virology 78, 6649–6656 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortola E, Noad R & Roy P Bluetongue virus outer capsid proteins are sufficient to trigger apoptosis in mammalian cells. Journal of Virology 78, 2875–2883 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall TM Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 15, 367–73 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Laity JH, Lee BM & Wright PE Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 11, 39–46 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Attar N et al. The Histone H3-H4 Tetramer is a Copper Reductase Enzyme. bioRxiv (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Jiang J, Sun J & Zhou ZH A putative ATPase mediates RNA transcription and capping in a dsRNA virus. Elife 4, e07901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carragher B et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J Struct Biol 132, 33–45 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ge P & Zhou ZH Hydrogen-bonding networks and RNA bases revealed by cryo electron microscopy suggest a triggering mechanism for calcium switches. Proc Natl Acad Sci U S A 108, 9637–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Jin L, Fang Q, Hui WH & Zhou ZH 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludtke SJ, Baldwin PR & Chiu W EMAN: Semi-automated software for high resolution single particle reconstructions. J Struct Biol 128, 82–97 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Mindell JA & Grigorieff N Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142, 334–47 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Diaz R, Rice WJ & Stokes DL Fourier-Bessel reconstruction of helical assemblies. Methods Enzymol 482, 131–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egelman EH The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol 157, 83–94 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Ge P et al. Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states. Nat Struct Mol Biol 22, 377–82 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clemens DL, Ge P, Lee BY, Horwitz MA & Zhou ZH Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartesaghi A et al. 2.2 A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science 348, 1147–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJ The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.