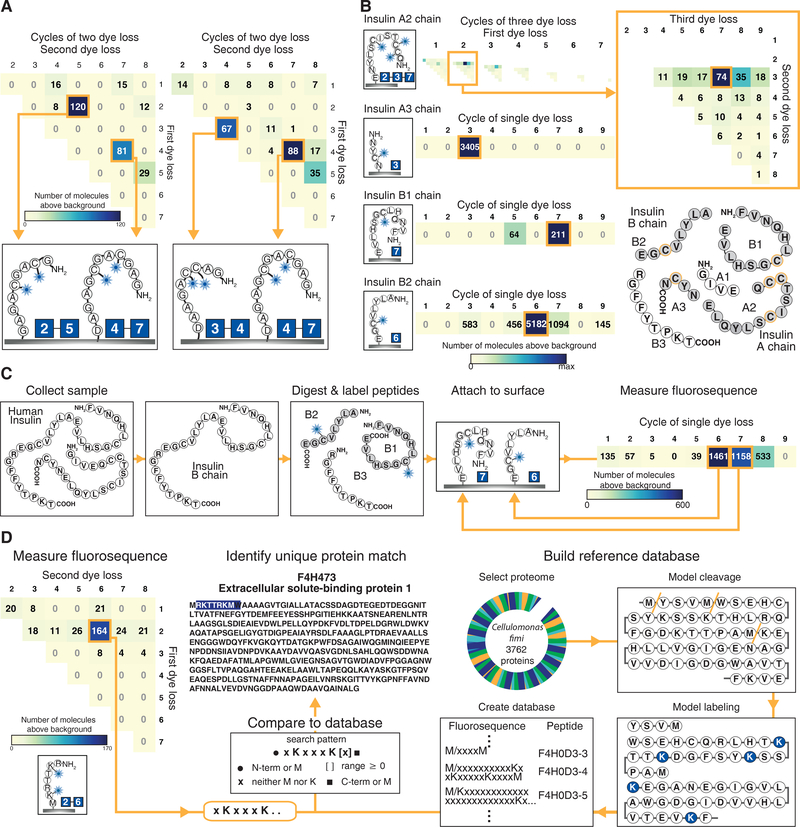
Figure 5: Fluorosequencing can discriminate individual peptide molecules in zeptomole-scale mixtures and uniquely identify their parent proteins.
(A) Histograms tallying counts of molecules sequenced from a mixture of GC♦AGC♦AGAG with GAGC♦GAC♦GAGAD (left panel, 98 image fields) and GAC♦C♦AGAAD with GAGC♦GAC♦GAGAD (right panel, 49 image fields) highlight the ability to distinguish individual peptides within mixtures. (B) Data on 4 individual insulin peptides that, in combination, uniquely identify insulin in the human proteome. (Top panel) adjusted three dye histogram for insulin A2 chain (QC♦C♦TSIC♦SLYNE) showing the expected signal at amino acid positions 2, 3, and 7 (magnified in inset). The remaining panels plot adjusted single dye histograms for insulin A3 (NYC♦N), B1 (FVNQHLC♦GSHLVE), and B2 chains (ALYLVC♦GE), respectively; each histogram represents 100 image fields. (C) Data on recombinant human insulin B chain after purification, GluC proteolysis and cysteine labeling shows the expected peaks at cycles 6 and 7 (100 image fields) as expected for the mixture of B2 and B1 peptides, respectively. (D) The fluorescent sequence of peptide RK†TTRK†M is sufficient to uniquely identify its parent protein F4H473 from the Cellulomonas fimi protein. The adjusted two dye sequencing histogram (left panel, 49 image fields) reveals the sequence as xKxxxK[x]≥0 which can be compared to a reference database (center panel) created by modeling fluorescent sequences for all possible peptides in the proteome assuming predefined protease cleavage and dye labeling specificities (right panel), here modeling cyanogen bromide cleavage after M and labeling K. ♦ indicates Atto647N conjugated to cysteine and † indicates Atto647N coupled to lysine residues. Supplementary Figs. 13–14 provide full single, double, and triple dye histograms, as appropriate.