Abstract
Early-life stressful experiences are critical for plasticity and development, shaping adult neuroendocrine response and future health. Stress response is mediated by the autonomous nervous system and the hypothalamic–pituitary–adrenal (HPA) axis while various environmental stimuli are encoded via epigenetic marks. The stress response system maintains homeostasis by regulating adaptation to the environmental changes. Pre-conceptual and in utero stressors form the fetal epigenetic profile together with the individual genetic profile, providing the background for individual stress response, vulnerability, or resilience. Postnatal and adult stressful experiences may act as the definitive switch. This review addresses the issue of how preconceptual in utero and postnatal events, together with individual differences, shape future stress responses. Putative markers of early-life adverse effects such as prematurity and low birth weight are emphasized, and the epigenetic, mitochondrial, and genomic architecture regulation of such events are discussed.
Keywords: stress, predisposition, epigenetics, low birth weight, individuality, early-life stress, mitochondria
Stress, Brain, and The Environment
Physiological or biological stress is the response to a stressor, i.e., an environmental condition or a stimulus. Τhe body responds to stress by sympathetic nervous system activation as a result of the fight-or-flight response. The stress response aims to restore homeostatic control and facilitate adaptation. The brain processes stress in three main areas: amygdala, hippocampus, and prefrontal cortex (PFC). Amygdala and hippocampus play a critical role in memory formation and are associated with anxiety, fear, and cognitive processes. PFC is the brain region linked to planning complex cognitive behavior, personality expression, decision making, and moderating social behavior (1). The basic activity of the PFC region is to orchestrate thoughts and actions in accordance with internal goals and executive function (2). Corticosteroid receptors that react to the stressor through steroid hormone binding are abundant in these areas (3, 4). It is well established that stressful experiences during critical periods of early brain development can affect emotional and behavioral functions in adult life (5). The autonomous nervous system and the hypothalamic–pituitary–adrenal (HPA) axis are responsible for these functions and mediate stress response through targeted hormone release. This system acts by negative feedback to maintain brain homeostasis. The hypothalamus is stimulated by its inputs and releases the corticotropin-releasing hormone. This hormone is transported to its target, the pituitary gland, where it binds to the targeted receptors and causes the release of the adrenocorticotropic hormone. Although the main purpose of this system is well understood, recent studies attempt to identify underlying genetic mechanisms of brain function modulating mediators of this system including adrenaline and neuropeptides (6). Glucocorticoids reach the brain through the peripheral blood flow, where they bind to specific types of cytoplasmic glucocorticoid receptors (GRs) and mineralocorticoid receptors (MRs). MRs make up the majority of stress corticosteroid receptors with a high affinity for cortisol and are activated as soon as a stressor appears. GRs have a low affinity for cortisol and are only activated when stress reaches its peak on the brain. This complex is then translocated to the nucleus, where it binds to specific DNA elements [glucocorticoid response elements (GREs)] and acts as a transcription factor activating or repressing a great number of genes (7).
Early-Life Stress, Learning, and Memory
Exposure to early-life stressful events has been shown to activate the HPA stress hormone system. HPA axis mediator and receptor genes are prime targets of epigenetic modifications by DNA methylation and histone acetylation (8). The combination of genetic and epigenetic factors affects cell function and brain development. As a result, individuals who have experienced chronic stress during early development and childhood are at high risk for a wide range of behavioral problems that persist into adulthood. This phenotype becomes evident by learning and emotion regulation difficulties, alcohol and substance abuse, externalizing problems, as well as depression and anxiety disorders (7). Children who have experienced maltreatment or were exposed to maternal deprivation trauma have shown poor performance in tasks involving working memory, attention, planning, and learning processes (9, 10). In rodents, maternal deprivation is a well-established paradigm of early-life stress. Maternal deprivation of newborns from their dam leads to epigenetic changes in specific imprinted genes and dysfunctions. Behavioral and molecular effects depend on the duration and type of maternal deprivation and individual predisposition (11).
In Utero Stress Exposures
Intrauterine life events may have a much greater impact on epigenetic profiles than stressful exposures during adult life (12). Early stages of embryonic development are characterized by heightened brain plasticity that is adversely affected by exposure to environmental insults (13). Complex gene environment interactions during critical early developmental periods may have lasting effects and result in adult psychopathology (14, 15). Maternal stress exposure, anxiety, and depression during pregnancy are considered in utero adverse experiences and have been associated with low birth weight (LBW) and future health problems (16–24). LBW, apart from being a risk factor for neonatal morbidity and mortality, has been proposed as a marker of early-life adversities (25, 26).
In this mini-review, genetic and epigenetic factors that shape stress response are discussed. The contribution of mitochondria and individual predisposition to developing mental health problems in response to a stressful stimulus will also be addressed.
Genetics and Epigenetics of The Stress Response
Early-life adversities have been implicated in the occurrence of neuropsychiatric conditions, such as, Post-traumatic stress disorder (PTSD), depression, psychosis, and phenotypes resembling mood- and anxiety-related disorders (4–8). Recent data are beginning to unravel the complex interactions between genes and environment, namely, an individual’s genetic and epigenetic profile that renders the person resilient or at risk for developing a stress-related disorder (9, 10). Apart from the genetics of neuroendocrine stress response, it is important to take into consideration its epigenetic profile (11). A plethora of epigenetic marks, contributing to either the enhanced or suppressed expression of a gene, in combination with risk- or resilience-related predisposing polymorphisms, shape an individual’s phenotype (27). The complex interaction of the genetic background with the epigenetic profile that reflects early-life experiences and is potentially reversible by environmental factors can result in a phenotype that is either resilient or sensitive towards adverse stress exposures (28, 29). Several genes and their epigenetic regulation have been implicated in the susceptibility to early-life stress. An overview of the below-discussed genes and their interrelations is provided in Figure 1 .
Figure 1.
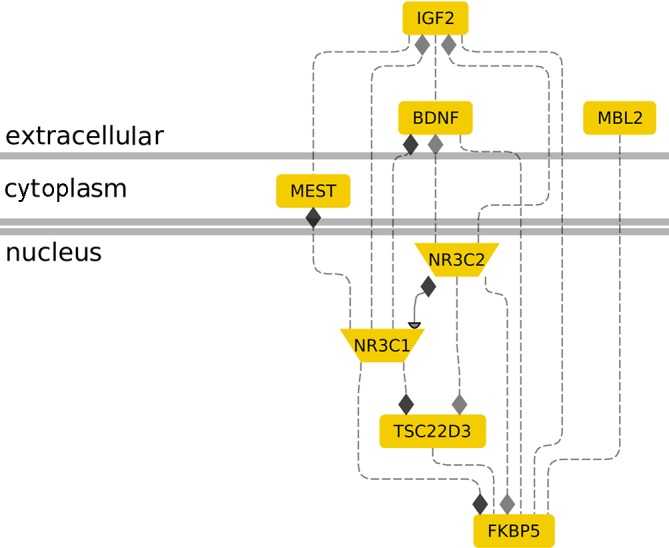
The role of molecular genetic markers in cellular stress response. The interconnection between genes IGF2, MBL2, MEST, NR3C1, NR3C2, TSC22D3, and BDNF in the context of stress response is shown. Membrane-bound IGF2 induction by maternal distress is associated with nuclear NR3C2 induction (3). IGF2 shows similar changes in methylation levels with NR3C1 during age-related stress (4), FKBP5 during the development of preterm infants (30), and MEST in infertile males (31). MBL2 and FKBP5 upregulation has been associated with parental-nutrition-induced stress (32, 33). NR3C2 and NR3C1 interact to control gene expression during stress (8, 34). Nuclear NR3C1 (glucocorticoid receptor) seems to be a convergence point for FKBP5 (9, 35), NR3C2 (10, 36), MEST (11, 37), and IGF2 (4) action in stress. TSC22D3 is an established glucocorticoid signaling responsive gene that is regulated by NR3C1 (27, 38) and an NR3C2 target (28). BDNF is upregulated by IGF2 (39) in an Alzheimer’s disease mouse model, inhibited by NR3C1 (40) in neuron-like cells, and associated with high NR3C2 and low NR3C1 in high-cholesterol-diet rats (41). FKBP5 elevation is associated with BDNF suppression and improved anxiety, depression, and posttraumatic stress disorder conditions (42). Gene interaction analysis was performed using the Genomatix Pathway System (Genomatix.de).
NR3C1 and NR3C2 genes (Nuclear Receptor Subfamily 3 Group C Member 1 and 2), encoding the GR and MR, respectively, are widely expressed in limbic regions of the brain and regulate HPA axis activity by cortisol binding. Deregulation of the GR–MR function may lead to HPA axis malfunction and stress vulnerability (43–45). The NR3C1 gene, localized on the 5q31-32 chromosome, contains nine exons (1–9) (45, 46). In the 5′ Untranslated region (UTR), alternative splice variants of the first exon form the distal and proximal gene promoter that contains a crucial CpG island regulating the expression of exon 1F. The multiple alternative first exon splice variants render the expression of NR3C1 tissue-specific (47–50). The first study in humans examining the epigenetic status of 1F promoter in low prenatal and increased maternal postnatal depression showed elevated methylation levels. This effect is reversed by maternal stroking of the newborns during the first postnatal weeks (51). In a thorough meta-analysis, psychosocial maternal prenatal stress was significantly correlated with DNA methylation at CpG 36 of the 1F promoter (52). Interestingly, prenatal exposure of depressed mothers to serotonin reuptake inhibitors was not associated with alterations in the methylation profile of the 1F promoter. However, a correlation between the psychological profiles of depressed mothers, especially during the third trimester, and increased HPA axis reactivity of the newborns, has been reported (53). Maternal anxiety during the first two trimesters also affects the methylation status of NR3C1, thus diminishing NR3C1 gene expression (54). In a study examining the effects of maternal-related stressors such as maternal deprivation due to financial difficulties, daily psychosocial stress, and war-related phenomena, a strong correlation was found between the aforementioned maternal stressors, neonatal birth weight, and methylation of multiple CpG sites in the upstream NR3C1 promoter. These results support the hypothesis that intrauterine development and maternal environmental stressors affect the plasticity and adaptation to adverse stimuli (55). Further supporting this notion, decreased expression of NR3C1 was observed in hippocampal tissues of suicide completers abused during childhood. These findings can be explained by alterations in hippocampal methylation of tissue-specific NR3C1, which persist into adulthood and lead to changes in HPA axis function (56–58). The NR3C2 gene on 4q31.1 has recently been associated with behavioral abnormalities. Cognitive ability following acute stress has been associated with genetic variation of the GR–MR. Specifically, single-nucleotide polymorphisms (SNPs) of the above genes seem to affect cognition and HPA axis function (59, 60). In individuals with a history of childhood maltreatment, the minor NR3C2 allele rs17581262 was correlated, among others, with lower amygdala and hippocampal volumes and major depression, suggesting that this allele is a predisposing risk factor for stress-related disorders (61).
FKBP5 (6p21.31) encodes a 51-kDa immunophilin, which is a major component of the GR heterocomplex. Upon stress exposure, cortisol diffuses into the cytoplasm and binds the GR (62–64). FKBP5 slows down the translocation of GR to the nucleus (65, 66). FKBP5 expression is regulated by GREs via a cortisol-dependent short negative feedback loop (67, 68). Ιn intron 2 of FKBP5 and close to a functional GRE, the significant SNP rs1360780 was identified (69). Structurally, the rare risk allele alters the chromatin conformation after GR binding to the GRE, inducing the transcription of FKBP5. In the presence of the protective allele, this induction is absent (67, 69). The aforementioned SNP has been linked to a variety of mental health conditions including depression, anxiety, psychosis, and posttraumatic stress disorder (70–72). During their in utero formation, brain regions including the amygdala and hippocampus are particularly vulnerable in cases of antenatal maternal depression and anxiety (73, 74). FKBP5 genetic variation among neonates combined with antenatal maternal depression can predispose toward the development of depressive symptoms in the offspring later in life due to alterations in neonatal brain regions (75). Interestingly, recent reports on the association of depression with childhood maltreatment did not report FKBP5 methylation to be involved in mediatory mechanisms (76, 77).
Alterations in GR function through NR3C1 lead to a rare endocrinological condition known as Primary Generalized Glucocorticoid Resistance (PGGR, Chrousos syndrome) (78, 79). Mutations in the NR3C1 gene result in receptor conformation changes and low ligand binding affinity and contribute to the clinical profile and pathogenesis (80–83). PGGR is characterized by decreased tissue sensitivity toward cortisol, resulting in malfunctioning negative feedback loops (84, 85). This causes a compensatory activation of the HPA axis and hypersecretion of its end products (80, 85, 86). Interestingly, FKBP5 has been implicated in glucocorticoid resistance. The gene’s overexpression is considered to be responsible for the low ligand-binding affinity of the GR in New World primates, providing a selective advantage of an overall normal adrenal function but with high concentrations of circulating Adrenocorticotropic hormone (ACTH) and cortisol (87, 88).
Brain-derived neurotrophic factor (BDNF) is a neurotrophin expressed in hippocampus and PFC affecting neuron survival, development, and plasticity. Early-life stress and Val66Met polymorphism result in lower BDNF availability (29, 89).
Τhe GILZ (glucocorticoid-induced leucine zipper) or TSC22D3 gene, located on Xq22.2 (90), is induced by cortisol-bound GR. This complex binds on the GRE in the promoter of GILZ, thus rendering this gene a valid measure of GR function (91–93). In an avian species, GILZ expression in the pituitary gland seems to be upregulated by glucocorticoids during the second half of the embryonic development and possibly plays a role in regulating pituitary hormone expression levels (94). GILZ is widely expressed in the brain, and its function depends on HPA axis activation. Increased expression of GILZ was found in the hippocampus and medial PFC of stressed mice, indicating a region-specific function (95). In human studies, decreased GILZ Messenger RNA (mRNA) levels were found in the PFC and the amygdaloid nuclei in teenage suicide completers (96). The above findings are only beginning to decipher the role of GILZ both in stress regulation and in immune system function.
Genetics and Epigenetics of Early Embryonic Development
MBL2 (mannose binding lectin 2) is an important regulator of innate immunity and inflammatory processes. The MBL2 gene encodes for a protein that assembles into a mannose-binding lectin complex. MBL2 plays a very important role in the first-line immune responses, as a component of neonate immunity when the adaptive immunity system is not sufficiently developed (97). In humans, MBL2 expression levels are determined genetically by a number of polymorphic sites of the gene as well as in its promoter region. Three non-synonymous SNPs, which are linked to absence or low levels of MBL2, have been identified in exon 1 and the promoter region. The most important MBL2 gene SNPs associated with early infection and preterm delivery risk are variants B [rs1800450 (GGC→GAC)], C [rs1800451 (GGA→GAA)], and D [rs5030737 (CGT→TGT)]. Moreover, there are SNPs in the promoter region at position −550 in variant H/L (rs11003125) and at position −221 in variant X/Y (rs7096206) (25, 98). These MBL2 gene polymorphisms are associated with an increased risk of perinatal and neonatal infections and risk of premature delivery (99, 100). MBL2 levels could not predict the risk of newborn morbidity or mortality as a single factor since morbidity is also affected by other factors including sex, premature delivery, birth weight, etc. (97).
IGF2 (insulin growth factor 2), an imprinted gene, acts as a growth factor promoting differentiation and metabolism and plays an important role in the development and nutritional needs of the fetus (101). IGF2 and H19 are two genes of the same imprinted domain expressed from the paternal and maternal allele, respectively, that have been implicated in the control of placental and embryonic growth through cell proliferation and apoptosis (102, 103). H19 is crucial for growth and differentiation of the placenta (104, 105).
MEST (mesoderm specific transcript, 7q32) is a paternally expressed imprinted gene, which influences placental and embryonic growth, as well as birth weight of the infant (31, 106). MEST is a member of the a/b-hydrolase superfamily and expressed in the embryonic mesoderm (107). Increased MEST expression is linked to infants with high birth weight. Decreased MEST gene expression is observed in premature embryos compared to normal embryos, but does not affect DNA methylation (108).
Maternal Stress and Mitochondria
Moving from single genes to subcellular functional systems, converging lines of evidence have pointed to an important role of mitochondria, the traditional “powerhouses of the cell,” as regulators of the stress response (109–111). Given the maternal origin and inheritance of mitochondria, it is plausible that maternal stress may exercise its effects on the offspring via alterations of mitochondrial pathways in both the in utero maternal microenvironment and offspring. Along these lines, it has been shown that maternal prenatal stress affects mitochondrial protein expression in pathways related to mitochondrial biogenesis and energy production in PFC and hippocampus of male rat offspring (112). Early-life maternal deprivation leads to a decrease in mitochondrial-related muscle gene expression in adult rats. Interestingly, adult-onset chronic stress had no effect on mitochondrial-related muscle gene expression function, indicating an early-life stress-specific effect (113). In humans, maternal psychosocial stress has been reported to alter the expression of mitochondrial proteins in the placenta (114). In this study, a link between mitochondrial changes and infant temperament has also been suggested. Maternal psychosocial stress and lifetime trauma have been associated with decreased mitochondrial DNA copy number in the placenta (115, 116).
Individuality
Chronic stress links changes in the epigenetic landscape with health conditions (117). Different cell types are characterized by distinct patterns of gene expression due to developmental, environmental, physiological, and pathological reasons (117). Epigenetic mechanisms affect gene function in a dynamic way as a result of different environmental exposures during fetal development. Early-life stressful experiences, such as nutritional deprivation, lack of maternal care, or chemical exposure during critical developmental periods, can lead to phenotypic differences later in life (118). In addition to genetic susceptibility (polymorphisms, genomic architecture) inter-individual phenotypic variations are also the result of epigenetic modifications. Once we realize how different environmental triggers affect the individual epigenetic processes, we may be able to develop new means to prevent or reverse environmentally driven epigenetic changes. A recent study supports this theory and suggests that adaptation to stress is a combination of three important factors: genetic predisposition, early-life environment, and late-life environment (119). In animal models, strain, age, sex, frequency, and duration of the stressor, time point within the light cycle and temperature, and even the housing conditions are some of the environmental factors that shape the stress response(120–122). In humans, genetic background, age, sex, type, frequency, and duration of the stressor and developmental stage have been suggested to be important factors that shape individual stress response (123).
Discussions Perspectives
Early-life stress can influence brain plasticity with lasting effects. Epigenetic factors including type of exposure, timing, and diversity of experience in combination with genetic predisposition contribute to the individual resilience or vulnerability toward stress. Elucidating the interplay and downstream affected pathways ( Figure 2 ) among i) housekeeping genes of the reproductive system, ii) regulators of the HPA axis, iii) components of mitochondrial heterogeneity, and iv) individual genomic architecture will facilitate our understanding of the impact of early-life stressful events for later life outcomes. Our analysis reveals the top 20 “satellite” genes ( Figure 2 ) that form a functional network, affecting and being affected by the core genes controlling early-life stress. Potentially stressful or compensatory individual experiences during lifetime may have an impact on the epigenetic landscape, thus masking the effects of early-life experiences. An improved understanding will allow an integrated, systemic approach to address pathological stress responses and pinpoint novel molecular targets for pharmacological and therapeutic interventions.
Figure 2.
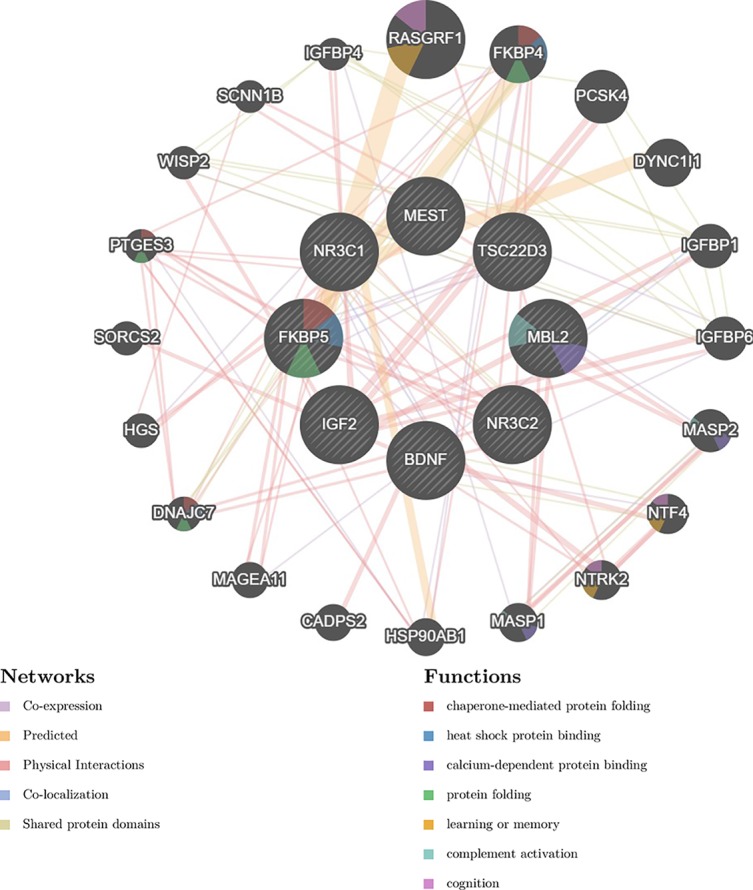
Gene interaction network discussed in the current review. The network was generated by the GeneMANIA prediction server (124). The left panel presents the different types of interactions with respective color coding, depicted with lines connecting genes in the network: physical interactions (pink), predicted (orange), co-expression (purple), and shared protein domains. The right panel presents gene functions, depicted with colored slices inside the respective genes: chaperone-mediated protein folding (red), heat shock protein binding (blue), calcium dependent protein binding (dark blue), protein folding (green), learning of memory (orange), complement activation (light blue), cognition (purple).
Author Contributions
All authors contributed to the writing and editing of the manuscript.
Funding
This work has received funding from the Hellenic Foundation for Research and Innovation (HFRI) and the General Secretariat for Research and Technology (GSRT), under grant agreement No 660.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Yang Y, Raine A. Prefrontal structural and functional brain imaging findings in antisocial, violent, and psychopathic individuals: a meta-analysis. Psychiatry Res (2009) 174(2):81–8. 10.1016/j.pscychresns.2009.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miller EK, Freedman DJ, Wallis JD. The prefrontal cortex: categories, concepts and cognition. Philos Trans R Soc Lond B Biol Sci (2002) 357(1424):1123–36. 10.1098/rstb.2002.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci (2005) 6(6):463–75. 10.1038/nrn1683 [DOI] [PubMed] [Google Scholar]
- 4. McEwen BS. Brain on stress: how the social environment gets under the skin. Proc Natl Acad Sci U S A (2012) 109 Suppl 2:17180–5. 10.1073/pnas.1121254109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor SE. Mechanisms linking early life stress to adult health outcomes. Proc Natl Acad Sci U S A (2010) 107(19):8507–12. 10.1073/pnas.1003890107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maras PM, Baram TZ. Sculpting the hippocampus from within: stress, spines, and CRH. Trends Neurosci (2012) 35(5):315–24. 10.1016/j.tins.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binder EB. Dissecting the molecular mechanisms of gene x environment interactions: implications for diagnosis and treatment of stress-related psychiatric disorders. Eur Jo Psychotraumatol (2017) 8(sup5):1412745. 10.1080/20008198.2017.1412745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murgatroyd C, Spengler D. Genetic variation in the epigenetic machinery and mental health. Curr Psychiatry Rep (2012) 14(2):138–49. 10.1007/s11920-012-0255-1 [DOI] [PubMed] [Google Scholar]
- 9. DePrince AP, Weinzierl KM, Combs MD. Executive function performance and trauma exposure in a community sample of children. Child Abuse Negl (2009) 33(6):353–61. 10.1016/j.chiabu.2008.08.002 [DOI] [PubMed] [Google Scholar]
- 10. Pollak SD. Multilevel developmental approaches to understanding the effects of child maltreatment: recent advances and future challenges. Dev Psychopathol (2015) 27(4 Pt 2):1387–97. 10.1017/S0954579415000826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwak HR, Lee JW, Kwon KJ, Kang CD, Cheong IY, Chun W, et al. Maternal social separation of adolescent rats induces hyperactivity and anxiolytic behavior. Korean J Physiol Pharmacol (2009) 13(2):79–83. 10.4196/kjpp.2009.13.2.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. King K, Murphy S, Hoyo C. Epigenetic regulation of Newborns’ imprinted genes related to gestational growth: patterning by parental race/ethnicity and maternal socioeconomic status. J Epidemiol Community Health (2015) 69(7):639–47. 10.1136/jech-2014-204781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green BB, Kappil M, Lambertini L, Armstrong DA, Guerin DJ, Sharp AJ, et al. Expression of imprinted genes in placenta is associated with infant neurobehavioral development. Epigenetics (2015) 10(9):834–41. 10.1080/15592294.2015.1073880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sale A, Berardi N, Maffei L. Environment and brain plasticity: towards an endogenous pharmacotherapy. Physiol Rev (2014) 94(1):189–234. 10.1152/physrev.00036.2012 [DOI] [PubMed] [Google Scholar]
- 15. Hartman S, Belsky J. Prenatal programming of postnatal plasticity revisited-and extended. Dev Psychopathol (2018) 30(3):825–42. 10.1017/S0954579418000548 [DOI] [PubMed] [Google Scholar]
- 16. Class QA, Lichtenstein P, Langstrom N, D’Onofrio BM. Timing of prenatal maternal exposure to severe life events and adverse pregnancy outcomes: a population study of 2.6 million pregnancies. Psychosom Med (2011) 73(3):234–41. 10.1097/PSY.0b013e31820a62ce [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Van den Bergh BR, Mulder EJ, Mennes M, Glover V. Antenatal maternal anxiety and stress and the neurobehavioural development of the fetus and child: links and possible mechanisms. Neurosci Biobehav Rev (2005) 29(2):237–58. 10.1016/j.neubiorev.2004.10.007 [DOI] [PubMed] [Google Scholar]
- 18. Slykerman RF, Thompson J, Waldie K, Murphy R, Wall C, Mitchell EA. Maternal stress during pregnancy is associated with moderate to severe depression in 11-year-old children. Acta Paediatr (2015) 104(1):68–74. 10.1111/apa.12787 [DOI] [PubMed] [Google Scholar]
- 19. Rondo PH, Ferreira RF, Nogueira F, Ribeiro MC, Lobert H, Artes R. Maternal psychological stress and distress as predictors of low birth weight, prematurity and intrauterine growth retardation. Eur J Clin Nutr (2003) 57(2):266–72. 10.1038/sj.ejcn.1601526 [DOI] [PubMed] [Google Scholar]
- 20. Mansell T, Novakovic B, Meyer B, Rzehak P, Vuillermin P, Ponsonby AL, et al. The effects of maternal anxiety during pregnancy on IGF2/H19 methylation in cord blood. Transl Psychiatry (2016) 6:e765. 10.1038/tp.2016.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janssen AB, Capron LE, O’Donnell K, Tunster SJ, Ramchandani PG, Heazell AE, et al. Maternal prenatal depression is associated with decreased placental expression of the imprinted gene PEG3. Psychol Med (2016) 46(14):2999–3011. 10.1017/S0033291716001598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tegethoff M, Greene N, Olsen J, Meyer AH, Meinlschmidt G. Maternal psychosocial adversity during pregnancy is associated with length of gestation and offspring size at birth: evidence from a population-based cohort study. Psychosom Med (2010) 72(4):419–26. 10.1097/PSY.0b013e3181d2f0b0 [DOI] [PubMed] [Google Scholar]
- 23. Grote NK, Bridge JA, Gavin AR, Melville JL, Iyengar S, Katon WJ. A meta-analysis of depression during pregnancy and the risk of preterm birth, low birth weight, and intrauterine growth restriction. Arch Gen Psychiatry (2010) 67(10):1012–24. 10.1001/archgenpsychiatry.2010.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grigoriadis S, VonderPorten EH, Mamisashvili L, Tomlinson G, Dennis CL, Koren G, et al. The impact of maternal depression during pregnancy on perinatal outcomes: a systematic review and meta-analysis. J Clin Psychiatry (2013) 74(4):e321–41. 10.4088/JCP.12r07968 [DOI] [PubMed] [Google Scholar]
- 25. Koroglu OA, Onay H, Erdemir G, Yalaz M, Cakmak B, Akisu M, et al. Mannose-binding lectin gene polymorphism and early neonatal outcome in preterm infants. Neonatology (2010) 98(4):305–12. 10.1159/000291487 [DOI] [PubMed] [Google Scholar]
- 26. Bodamer OA, Mitterer G, Maurer W, Pollak A, Mueller MW, Schmidt WM. Evidence for an association between mannose-binding lectin 2 (MBL2) gene polymorphisms and pre-term birth. Genet Med (2006) 8(8):518–24. 10.109701.gim.0000232478.43335.19 [DOI] [PubMed] [Google Scholar]
- 27. Madruga C, Xavier LL, Achaval M, Sanvitto GL, Lucion AB. Early handling, but not maternal separation, decreases emotional responses in two paradigms of fear without changes in mesolimbic dopamine. Behav Brain Res (2006) 166(2):241–6. 10.1016/j.bbr.2005.08.005 [DOI] [PubMed] [Google Scholar]
- 28. Lee DT, Yip AS, Chiu HF, Leung TY, Chung TK. Screening for postnatal depression: are specific instruments mandatory? J Affect Disord (2001) 63(1–3):233–8. 10.1016/S0165-0327(00)00193-2 [DOI] [PubMed] [Google Scholar]
- 29. Casavant SG, Cong X, Fitch RH, Moore J, Rosenkrantz T, Starkweather A. Allostatic load and biomarkers of stress in the preterm infant: an integrative review. Biol Res Nurs (2019) 21(2):210–28. 10.1177/1099800418824415 [DOI] [PubMed] [Google Scholar]
- 30. Taylor A, Littlewood J, Adams D, Dore C, Glover V. Serum cortisol levels are related to moods of elation and dysphoria in new mothers. Psychiatry Res (1994) 54(3):241–7. 10.1016/0165-1781(94)90018-3 [DOI] [PubMed] [Google Scholar]
- 31. Huntriss JD, Hemmings KE, Hinkins M, Rutherford AJ, Sturmey RG, Elder K, et al. Variable imprinting of the MEST gene in human preimplantation embryos. Eur J Hum Genet (2013) 21(1):40–7. 10.1038/ejhg.2012.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wildhaber BE, Yang H, Tazuke Y, Teitelbaum DH. Gene alteration of intestinal intraepithelial lymphocytes with administration of total parenteral nutrition. J Pediatr Surg (2003) 38(6):840–3. 10.1016/S0022-3468(03)00107-6 [DOI] [PubMed] [Google Scholar]
- 33. Binder EB. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology (2009) 34 Suppl 1:S186–95. 10.1016/j.psyneuen.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 34. Oitzl MS, de Kloet ER, Joels M, Schmid W, Cole TJ. Spatial learning deficits in mice with a targeted glucocorticoid receptor gene disruption. Eur J Neurosci (1997) 9(11):2284–96. 10.1111/j.1460-9568.1997.tb01646.x [DOI] [PubMed] [Google Scholar]
- 35. Tyrka AR, Ridout KK, Parade SH, Paquette A, Marsit CJ, Seifer R. Childhood maltreatment and methylation of FK506 binding protein 5 gene (FKBP5). Dev Psychopathol (2015) 27(4 Pt 2):1637–45. 10.1017/S0954579415000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spencer RL, Kim PJ, Kalman BA, Cole MA. Evidence for mineralocorticoid receptor facilitation of glucocorticoid receptor-dependent regulation of hypothalamic-pituitary-adrenal axis activity. Endocrinology (1998) 139(6):2718–26. 10.1210/endo.139.6.6029 [DOI] [PubMed] [Google Scholar]
- 37. El Hajj N, Pliushch G, Schneider E, Dittrich M, Muller T, Korenkov M, et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes (2013) 62(4):1320–8. 10.2337/db12-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol (2006) 20(3):560–72. 10.1210/me.2005-0318 [DOI] [PubMed] [Google Scholar]
- 39. Mellott TJ, Pender SM, Burke RM, Langley EA, Blusztajn JK. IGF2 ameliorates amyloidosis, increases cholinergic marker expression and raises BMP9 and neurotrophin levels in the hippocampus of the APPswePS1dE9 Alzheimer’s disease model mice. PLoS One (2014) 9(4):e94287. 10.1371/journal.pone.0094287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen H, Lombes M, Le Menuet D. Glucocorticoid receptor represses brain-derived neurotrophic factor expression in neuron-like cells. Mol Brain (2017) 10(1):12. 10.1186/s13041-017-0295-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu X, Wang T, Luo J, Liang S, Li W, Wu X, et al. Age-dependent effect of high cholesterol diets on anxiety-like behavior in elevated plus maze test in rats. Behav Brain Funct (2014) 10:30. 10.1186/1744-9081-10-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hammamieh R, Chakraborty N, Gautam A, Miller SA, Muhie S, Meyerhoff J, et al. Transcriptomic analysis of the effects of a fish oil enriched diet on murine brains. PLoS One (2014) 9(3):e90425. 10.1371/journal.pone.0090425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. DeRijk R, de Kloet ER. Corticosteroid receptor genetic polymorphisms and stress responsivity. Endocrine (2005) 28(3):263–70. 10.1385/ENDO:28:3:263 [DOI] [PubMed] [Google Scholar]
- 44. Funder JW. Mineralocorticoids, glucocorticoids, receptors and response elements. Science (New York, NY) (1993) 259(5098):1132–3. 10.1126/science.8382375 [DOI] [PubMed] [Google Scholar]
- 45. Kino T, Su YA, Chrousos GP. Human glucocorticoid receptor isoform beta: recent understanding of its potential implications in physiology and pathophysiology. Cell Mol Life Sci (2009) 66(21):3435–48. 10.1007/s00018-009-0098-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Petta I, Dejager L, Ballegeer M, Lievens S, Tavernier J, De Bosscher K, et al. The interactome of the glucocorticoid receptor and its influence on the actions of glucocorticoids in combatting inflammatory and infectious diseases. Microbiol Mol Biol Rev (2016) 80(2):495–522. 10.1128/MMBR.00064-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daskalakis NP, Yehuda R. Site-specific methylation changes in the glucocorticoid receptor exon 1F promoter in relation to life adversity: systematic review of contributing factors. Front Neurosci (2014) 8:369. 10.3389/fnins.2014.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turner JD, Muller CP. Structure of the glucocorticoid receptor (NR3C1) gene 5’ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol (2005) 35(2):283–92. 10.1677/jme.1.01822 [DOI] [PubMed] [Google Scholar]
- 49. Turner JD, Alt SR, Cao L, Vernocchi S, Trifonova S, Battello N, et al. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem Pharmacol (2010) 80(12):1860–8. 10.1016/j.bcp.2010.06.037 [DOI] [PubMed] [Google Scholar]
- 50. Vandevyver S, Dejager L, Libert C. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr Rev (2014) 35(4):671–93. 10.1210/er.2014-1010 [DOI] [PubMed] [Google Scholar]
- 51. Murgatroyd C, Quinn JP, Sharp HM, Pickles A, Hill J. Effects of prenatal and postnatal depression, and maternal stroking, at the glucocorticoid receptor gene. Transl Psychiatry (2015) 5:e560. 10.1038/tp.2014.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Palma-Gudiel H, Cordova-Palomera A, Eixarch E, Deuschle M, Fananas L. Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis. Epigenetics (2015) 10(10):893–902. 10.1080/15592294.2015.1088630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics (2008) 3(2):97–106. 10.4161/epi.3.2.6034 [DOI] [PubMed] [Google Scholar]
- 54. Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A, et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatr Res (2013) 47(7):880–91. 10.1016/j.jpsychires.2013.03.009 [DOI] [PubMed] [Google Scholar]
- 55. Mulligan CJ, D’Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics (2012) 7(8):853–7. 10.4161/epi.21180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McGowan PO, Sasaki A, D’Alessio AC, Dymov S, Labonte B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci (2009) 12(3):342–8. 10.1038/nn.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tyrka AR, Ridout KK, Parade SH. Childhood adversity and epigenetic regulation of glucocorticoid signaling genes: associations in children and adults. Dev Psychopathol (2016) 28(4pt2):1319–31. 10.1017/S0954579416000870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry (2011) 1:e59. 10.1038/tp.2011.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Keller J, Gomez R, Williams G, Lembke A, Lazzeroni L, Murphy GM, Jr., et al. HPA axis in major depression: cortisol, clinical symptomatology and genetic variation predict cognition. Mol Psychiatry (2017) 22(4):527–36. 10.1038/mp.2016.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Plieger T, Felten A, Splittgerber H, Duke E, Reuter M. The role of genetic variation in the glucocorticoid receptor (NR3C1) and mineralocorticoid receptor (NR3C2) in the association between cortisol response and cognition under acute stress. Psychoneuroendocrinology (2018) 87:173–80. 10.1016/j.psyneuen.2017.10.020 [DOI] [PubMed] [Google Scholar]
- 61. Gerritsen L, Milaneschi Y, Vinkers CH, van Hemert AM, van Velzen L, Schmaal L, et al. HPA axis genes, and their interaction with childhood maltreatment, are related to cortisol levels and stress-related phenotypes. Neuropsychopharmacology (2017) 42(12):2446–55. 10.1038/npp.2017.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wiederrecht G, Hung S, Chan HK, Marcy A, Martin M, Calaycay J, et al. Characterization of high molecular weight FK-506 binding activities reveals a novel FK-506-binding protein as well as a protein complex. J Biol Chem (1992) 267(30):21753–60. [PubMed] [Google Scholar]
- 63. Sanchez ER. Chaperoning steroidal physiology: lessons from mouse genetic models of Hsp90 and its cochaperones. Biochim Biophys Acta (2012) 1823(3):722–9. 10.1016/j.bbamcr.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pelleymounter LL, Moon I, Johnson JA, Laederach A, Halvorsen M, Eckloff B, et al. A novel application of pattern recognition for accurate SNP and indel discovery from high-throughput data: targeted resequencing of the glucocorticoid receptor co-chaperone FKBP5 in a Caucasian population. Mol Genet Metab (2011) 104(4):457–69. 10.1016/j.ymgme.2011.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wochnik GM, Ruegg J, Abel GA, Schmidt U, Holsboer F, Rein T. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem (2005) 280(6):4609–16. 10.1074/jbc.M407498200 [DOI] [PubMed] [Google Scholar]
- 66. Davies TH, Ning YM, Sanchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J Biol Chem (2002) 277(7):4597–600. 10.1074/jbc.C100531200 [DOI] [PubMed] [Google Scholar]
- 67. Paakinaho V, Makkonen H, Jaaskelainen T, Palvimo JJ. Glucocorticoid receptor activates poised FKBP51 locus through long-distance interactions. Mol Endocrinol (2010) 24(3):511–25. 10.1210/me.2009-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hubler TR, Scammell JG. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones (2004) 9(3):243–52. 10.1379/CSC-32R.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci (2013) 16(1):33–41. 10.1038/nn.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet (2004) 36(12):1319–25. 10.1038/ng1479 [DOI] [PubMed] [Google Scholar]
- 71. Criado-Marrero M, Rein T, Binder EB, Porter JT, Koren J, Blair LJ. Hsp90 and FKBP51: complex regulators of psychiatric diseases. Philos Trans R Soc Lond B Biol Sci (2018) 373, 1738. 10.1098/rstb.2016.0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Voisey J, Young RM, Lawford BR, Morris CP. Progress towards understanding the genetics of posttraumatic stress disorder. J Anxiety Disord (2014) 28(8):873–83. 10.1016/j.janxdis.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 73. Qiu A, Anh TT, Li Y, Chen H, Rifkin-Graboi A, Broekman BF, et al. Prenatal maternal depression alters amygdala functional connectivity in 6-month-old infants. Transl Psychiatry (2015) 5:e508. 10.1038/tp.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qiu A, Rifkin-Graboi A, Chen H, Chong YS, Kwek K, Gluckman PD, et al. Maternal anxiety and infants’ hippocampal development: timing matters. Transl Psychiatry (2013) 3:e306. 10.1038/tp.2013.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang C, Shen M, Guillaume B, Chong YS, Chen H, Fortier MV, et al. FKBP5 moderates the association between antenatal maternal depressive symptoms and neonatal brain morphology. Neuropsychopharmacology (2018) 43(3):564–70. 10.1038/npp.2017.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bustamante AC, Aiello AE, Guffanti G, Galea S, Wildman DE, Uddin M. FKBP5 DNA methylation does not mediate the association between childhood maltreatment and depression symptom severity in the Detroit Neighborhood Health Study. J Psychiatr Res (2018) 96:39–48. 10.1016/j.jpsychires.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. D’Agata AL, Walsh S, Vittner D, Cong X, McGrath JM, Young EE. FKBP5 genotype and early life stress exposure predict neurobehavioral outcomes for preterm infants. Dev Psychobiol (2017) 59(3):410–8. 10.1002/dev.21507 [DOI] [PubMed] [Google Scholar]
- 78. Charmandari E. Primary generalized glucocorticoid resistance and hypersensitivity. Horm Res Paediatr (2011) 76(3):145–55. 10.1159/000330759 [DOI] [PubMed] [Google Scholar]
- 79. Charmandari E. Primary generalized glucocorticoid resistance and hypersensitivity: the end-organ involvement in the stress response. Sci Signal (2012) 5(244). 10.1126/scisignal.2003337 [DOI] [PubMed] [Google Scholar]
- 80. Nicolaides N, Lamprokostopoulou A, Sertedaki A, Charmandari E. Recent advances in the molecular mechanisms causing primary generalized glucocorticoid resistance. Hormones (Athens) (2016) 15(1):23–34. 10.14310/horm.2002.1660 [DOI] [PubMed] [Google Scholar]
- 81. Nicolaides NC, Geer EB, Vlachakis D, Roberts ML, Psarra AM, Moutsatsou P, et al. A novel mutation of the hGR gene causing Chrousos syndrome. Eur J Clin Invest (2015) 45(8):782–91. 10.1111/eci.12470 [DOI] [PubMed] [Google Scholar]
- 82. Nicolaides NC, Skyrla E, Vlachakis D, Psarra AM, Moutsatsou P, Sertedaki A, et al. Functional characterization of the hGRalphaT556I causing Chrousos syndrome. Eur J Clin Invest (2016) 46(1):42–9. 10.1111/eci.12563 [DOI] [PubMed] [Google Scholar]
- 83. Hurt DE, Suzuki S, Mayama T, Charmandari E, Kino T. Structural analysis on the pathologic mutant glucocorticoid receptor ligand-binding domains. Mol Endocrinol (2016) 30(2):173–88. 10.1210/me.2015-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chrousos G. Q&A: primary generalized glucocorticoid resistance. BMC Med (2011) 9:27. 10.1186/1741-7015-9-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chrousos GP, Detera-Wadleigh SD, Karl M. Syndromes of glucocorticoid resistance. Ann Intern Med (1993) 119(11):1113–24. 10.7326/0003-4819-119-11-199312010-00009 [DOI] [PubMed] [Google Scholar]
- 86. Charmandari E, Kino T. Chrousos syndrome: a seminal report, a phylogenetic enigma and the clinical implications of glucocorticoid signalling changes. Eur J Clin Invest (2010) 40(10):932–42. 10.1111/j.1365-2362.2010.02336.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Reynolds PD, Ruan Y, Smith DF, Scammell JG. Glucocorticoid resistance in the squirrel monkey is associated with overexpression of the immunophilin FKBP51. J Clin Endocrinol Metab (1999) 84(2):663–9. 10.1210/jcem.84.2.5429 [DOI] [PubMed] [Google Scholar]
- 88. Scammell JG, Denny WB, Valentine DL, Smith DF. Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three New World primates. Gen Comp Endocrinol (2001) 124(2):152–65. 10.1006/gcen.2001.7696 [DOI] [PubMed] [Google Scholar]
- 89. Chau CM, Cepeda IL, Devlin AM, Weinberg J, Grunau RE. The Val66Met brain-derived neurotrophic factor gene variant interacts with early pain exposure to predict cortisol dysregulation in 7-year-old children born very preterm: implications for cognition. Neuroscience (2017) 342:188–99. 10.1016/j.neuroscience.2015.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Cannarile L, Zollo O, D’Adamio F, Ayroldi E, Marchetti C, Tabilio A, et al. Cloning, chromosomal assignment and tissue distribution of human GILZ, a glucocorticoid hormone-induced gene. Cell Death Differ (2001) 8(2):201–3. 10.1038/sj.cdd.4400798 [DOI] [PubMed] [Google Scholar]
- 91. D’Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, et al. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity (1997) 7(6):803–12. 10.1016/S1074-7613(00)80398-2 [DOI] [PubMed] [Google Scholar]
- 92. Thiagarajah AS, Eades LE, Thomas PR, Guymer EK, Morand EF, Clarke DM, et al. GILZ: glitzing up our understanding of the glucocorticoid receptor in psychopathology. Brain Res (2014) 1574:60–9. 10.1016/j.brainres.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 93. van der Laan S, Sarabdjitsingh RA, Van Batenburg MF, Lachize SB, Li H, Dijkmans TF, et al. Chromatin immunoprecipitation scanning identifies glucocorticoid receptor binding regions in the proximal promoter of a ubiquitously expressed glucocorticoid target gene in brain. J Neurochem (2008) 106(6):2515–23. 10.1111/j.1471-4159.2008.05575.x [DOI] [PubMed] [Google Scholar]
- 94. Ellestad LE, Malkiewicz SA, Guthrie HD, Welch GR, Porter TE. Expression and regulation of glucocorticoid-induced leucine zipper in the developing anterior pituitary gland. J Mol Endocrinol (2009) 42(2):171–83. 10.1677/JME-08-0066 [DOI] [PubMed] [Google Scholar]
- 95. Yachi K, Inoue K, Tanaka H, Yoshikawa H, Tohyama M. Localization of glucocorticoid-induced leucine zipper (GILZ) expressing neurons in the central nervous system and its relationship to the stress response. Brain Res (2007) 1159:141–7. 10.1016/j.brainres.2007.05.024 [DOI] [PubMed] [Google Scholar]
- 96. Pandey GN, Rizavi HS, Ren X, Dwivedi Y, Palkovits M. Region-specific alterations in glucocorticoid receptor expression in the postmortem brain of teenage suicide victims. Psychoneuroendocrinology (2013) 38(11):2628–39. 10.1016/j.psyneuen.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Speletas M, Gounaris A, Sevdali E, Kompoti M, Konstantinidi K, Sokou R, et al. MBL2 genotypes and their associations with MBL levels and NICU morbidity in a cohort of Greek neonates. J Immunol Res (2015) 2015:478412. 10.1155/2015/478412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Steffensen R, Hoffmann K, Varming K. Rapid genotyping of MBL2 gene mutations using real-time PCR with fluorescent hybridisation probes. J Immunol Methods (2003) 278(1–2):191–9. 10.1016/S0022-1759(03)00190-X [DOI] [PubMed] [Google Scholar]
- 99. Swierzko AS, Atkinson AP, Cedzynski M, Macdonald SL, Szala A, Domzalska-Popadiuk I, et al. Two factors of the lectin pathway of complement, l-ficolin and mannan-binding lectin, and their associations with prematurity, low birthweight and infections in a large cohort of Polish neonates. Mol Immunol (2009) 46(4):551–8. 10.1016/j.molimm.2008.07.025 [DOI] [PubMed] [Google Scholar]
- 100. Hilgendorff A, Heidinger K, Pfeiffer A, Bohnert A, Konig IR, Ziegler A, et al. Association of polymorphisms in the mannose-binding lectin gene and pulmonary morbidity in preterm infants. Genes Immun (2007) 8(8):671–7. 10.1038/sj.gene.6364432 [DOI] [PubMed] [Google Scholar]
- 101. Harris LK, Crocker IP, Baker PN, Aplin JD, Westwood M. IGF2 actions on trophoblast in human placenta are regulated by the insulin-like growth factor 2 receptor, which can function as both a signaling and clearance receptor. Biol Reprod (2011) 84(3):440–6. 10.1095/biolreprod.110.088195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Vidal AC, Henry NM, Murphy SK, Oneko O, Nye M, Bartlett JA, et al. PEG1/MEST and IGF2 DNA methylation in CIN and in cervical cancer. Clin Transl Oncol (2014) 16(3):266–72. 10.1007/s12094-013-1067-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Huang RC, Galati JC, Burrows S, Beilin LJ, Li X, Pennell CE, et al. DNA methylation of the IGF2/H19 imprinting control region and adiposity distribution in young adults. Clin Epigenetics (2012) 4(1):21. 10.1186/1868-7083-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yu L, Chen M, Zhao D, Yi P, Lu L, Han J, et al. The H19 gene imprinting in normal pregnancy and pre-eclampsia. Placenta (2009) 30(5):443–7. 10.1016/j.placenta.2009.02.011 [DOI] [PubMed] [Google Scholar]
- 105. Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, Jones A, et al. Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med (2011) 28(4):481–7. 10.3892/ijmm.2011.754 [DOI] [PubMed] [Google Scholar]
- 106. Zechner U, Pliushch G, Schneider E, El Hajj N, Tresch A, Shufaro Y, et al. Quantitative methylation analysis of developmentally important genes in human pregnancy losses after ART and spontaneous conception. Mol Hum Reprod (2010) 16(9):704–13. 10.1093/molehr/gap107 [DOI] [PubMed] [Google Scholar]
- 107. Vidal AC, Benjamin Neelon SE, Liu Y, Tuli AM, Fuemmeler BF, Hoyo C, et al. Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans. Genet Epigenet (2014) 6:37–44. 10.4137/GEG.S18067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta (2006) 27(6–7):540–9. 10.1016/j.placenta.2005.07.004 [DOI] [PubMed] [Google Scholar]
- 109. Picard M, McEwen BS, Epel ES, Sandi C. An energetic view of stress: focus on mitochondria. Front Neuroendocrinol (2018) 49:72–85. 10.1016/j.yfrne.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lopes S, Teplytska L, Vaz-Silva J, Dioli C, Trindade R, Morais M, et al. Tau deletion prevents stress-induced dendritic atrophy in prefrontal cortex: role of synaptic mitochondria. Cereb Cortex (2017) 27(4):2580–91. 10.1093/cercor/bhw057 [DOI] [PubMed] [Google Scholar]
- 111. Manoli I, Alesci S, Blackman MR, Su YA, Rennert OM, Chrousos GP. Mitochondria as key components of the stress response. Trends Endocrinol Metab (2007) 18(5):190–8. 10.1016/j.tem.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 112. Glombik K, Stachowicz A, Slusarczyk J, Trojan E, Budziszewska B, Suski M, et al. Maternal stress predicts altered biogenesis and the profile of mitochondrial proteins in the frontal cortex and hippocampus of adult offspring rats. Psychoneuroendocrinology (2015) 60:151–62. 10.1016/j.psyneuen.2015.06.015 [DOI] [PubMed] [Google Scholar]
- 113. Ghosh S, Banerjee KK, Vaidya VA, Kolthur-Seetharam U. Early stress history alters serum insulin-like growth factor-1 and impairs muscle mitochondrial function in adult male rats. J Neuroendocrinol (2016) 28(9). 10.1111/jne.12397 [DOI] [PubMed] [Google Scholar]
- 114. Lambertini L, Chen J, Nomura Y. Mitochondrial gene expression profiles are associated with maternal psychosocial stress in pregnancy and infant temperament. PLoS One (2015) 10(9):e0138929. 10.1371/journal.pone.0138929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Brunst KJ, Sanchez Guerra M, Gennings C, Hacker M, Jara C, Bosquet Enlow M, et al. Maternal lifetime stress and prenatal psychological functioning and decreased placental mitochondrial DNA copy number in the PRISM study. Am J Epidemiol (2017) 186(11):1227–36. 10.1093/aje/kwx183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Brunst KJ, Sanchez-Guerra M, Chiu YM, Wilson A, Coull BA, Kloog I, et al. Prenatal particulate matter exposure and mitochondrial dysfunction at the maternal-fetal interface: effect modification by maternal lifetime trauma and child sex. Environ Int (2018) 112:49–58. 10.1016/j.envint.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Johnstone SE, Baylin SB. Stress and the epigenetic landscape: a link to the pathobiology of human diseases? Nat Rev Genet (2010) 11(11):806–12. 10.1038/nrg2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reproductive Toxicol (2007) 24(1):9–19. 10.1016/j.reprotox.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 119. Daskalakis NP, Bagot RC, Parker KJ, Vinkers CH, de Kloet ER. The three-hit concept of vulnerability and resilience: toward understanding adaptation to early-life adversity outcome. Psychoneuroendocrinology (2013) 38(9):1858–73. 10.1016/j.psyneuen.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Richter SH, Garner JP, Wurbel H. Environmental standardization: cure or cause of poor reproducibility in animal experiments? Nat Methods (2009) 6(4):257–61. 10.1038/nmeth.1312 [DOI] [PubMed] [Google Scholar]
- 121. Tsai PP, Pachowsky U, Stelzer HD, Hackbarth H. Impact of environmental enrichment in mice. 1: effect of housing conditions on body weight, organ weights and haematology in different strains. Lab Anim (2002) 36(4):411–9. 10.1258/002367702320389071 [DOI] [PubMed] [Google Scholar]
- 122. Festing MF, Altman DG. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J (2002) 43(4):244–58. 10.1093/ilar.43.4.244 [DOI] [PubMed] [Google Scholar]
- 123. Bale TL. The placenta and neurodevelopment: sex differences in prenatal vulnerability. Dialogues Clin Neurosci (2016) 18(4):459–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res (2010) 38(Web Server issue):W214–20. 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]


