 Based on the scaffold of astemizole, three novel turn-on fluorescent probes (N1–N3) for human ether-a-go-go-related gene (hERG) potassium channel were developed herein.
Based on the scaffold of astemizole, three novel turn-on fluorescent probes (N1–N3) for human ether-a-go-go-related gene (hERG) potassium channel were developed herein.
Abstract
Based on the scaffold of astemizole, three novel turn-on fluorescent probes (N1–N3) for human ether-a-go-go-related gene (hERG) potassium channel were developed herein. These probes have reasonable fluorescence properties, acceptable cell toxicity, and potent inhibitory activity, all of which contribute to cell imaging at the nanomolar level. Overall, these probes have the potential for setting up a screening system for hERG channels.
Introduction
The human ether-a-go-go-related gene encodes hERG potassium channel (Kv11.1), a key effector of cardiac repolarization.1 hERG channel is a voltage-dependent channel known for its role in repolarizing the cardiac action potential. The alteration of hERG by mutation2 or pharmacological inhibition3 produces long QT syndrome and the lethal cardiac arrhythmia torsade de pointes. In recent years, more and more drugs have started to exhibit serious adverse cardiac reactions due to their blockade of the hERG channel. Most of them were withdrawn from the market, such as cisapride, tefenadine, astemizole and grepafloxacin.4 Currently, all drugs should be measured for their affinity towards hERG channel to evaluate their cardiotoxicity, which is required by the FDA. Recently, MacKinnon et al.5 discovered that hERG channel is opened by the voltage sensors when they are in a depolarized conformation and the hERG channel's idiosyncratic sensitivity to many drugs may be associated with the very small volume of the central cavity, which is embraced by four deep hydrophobic pockets. However, there is still no simple and effective method for a high-throughput screening of drugs with respect to the hERG channel.
Recently, many studies have shown that in comparison with the corresponding normal cells, the expression of hERG channel is up-regulated in some tumor cell lines6–8 such as neuroblastoma, breast cancer, and colon cancer cells to facilitate cell proliferation, invasion, and tumor angiogenesis.9 However, there is no explanation for the up-regulation of hERG channel and the role of hERG channel in these cancers. For the purpose of better analysis and molecular imaging of hERG channels, there is a stringent demand for a simple, cost-effective, and safe method to label hERG channels. This may help to explain the role of hERG channel in these cancers so as to make hERG channel become a biomarker and a new target for tumor research.
At the present time, small-molecule fluorescent probes have been diffusely used in pharmacology and biology for the detection of proteins, nucleic acids, and other vital biological molecules, owing to their unique properties, such as high sensitivity, simplicity, and convenient operation. When they label targets, they can provide real-time information and trace the dynamic process on cells even at the animal level with high spatio-temporal resolution.10–12
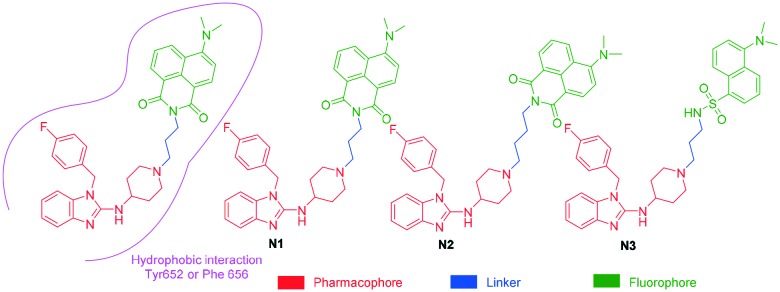
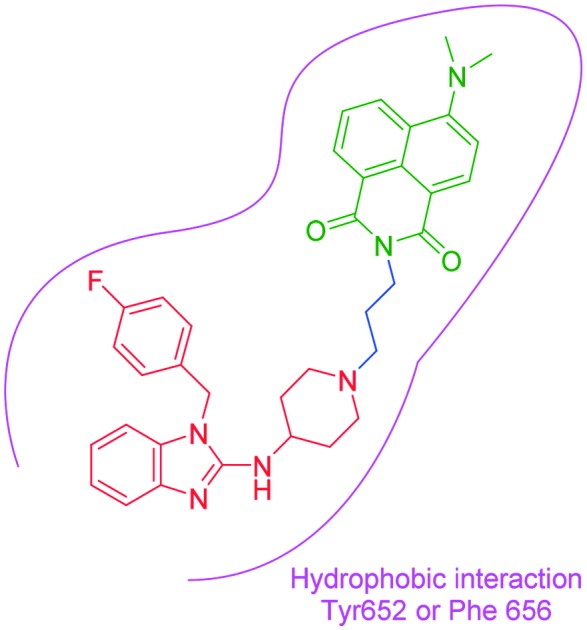
It has been well studied that when a ligand binds to the hERG channel, a hydrophobic interaction between the ligand and the residue Phe 656 or Tyr 652 of hERG channel plays a critical role in the high-affinity binding1,13,14 thus, we decided to use the molecular mechanism to design environment-sensitive probes. In fact, probes with the off–on mechanism have been utilized to detect structural characterization of ligand-binding domains, protein–protein interactions, and protein conformation dynamics.15–17 Generally, three parts are included in a typical fluorescent probe: a pharmacophore, a fluorophore, and a linker. Astemizole, the most potent inhibitor of the hERG channel, was chosen as the pharmacophore. In addition, some fluorophores with good fluorescent properties could be used in small-molecule fluorescent probes, such as naphthalimide18 and dansyl derivative.19 In order to keep the inhibitory activity of the probe, the major interaction sites of astemizole's bonding were retained. Herein, the naphthalimide and dansyl fluorophores were conjugated to the recognition moiety using an aliphatic spacer. Soon afterwards, several small-molecule fluorescent probes (N1–N3) for the hERG channel were synthesized (Scheme 1).
Scheme 1. Design strategy of the fluorescent probes.
Results and discussion
Chemistry
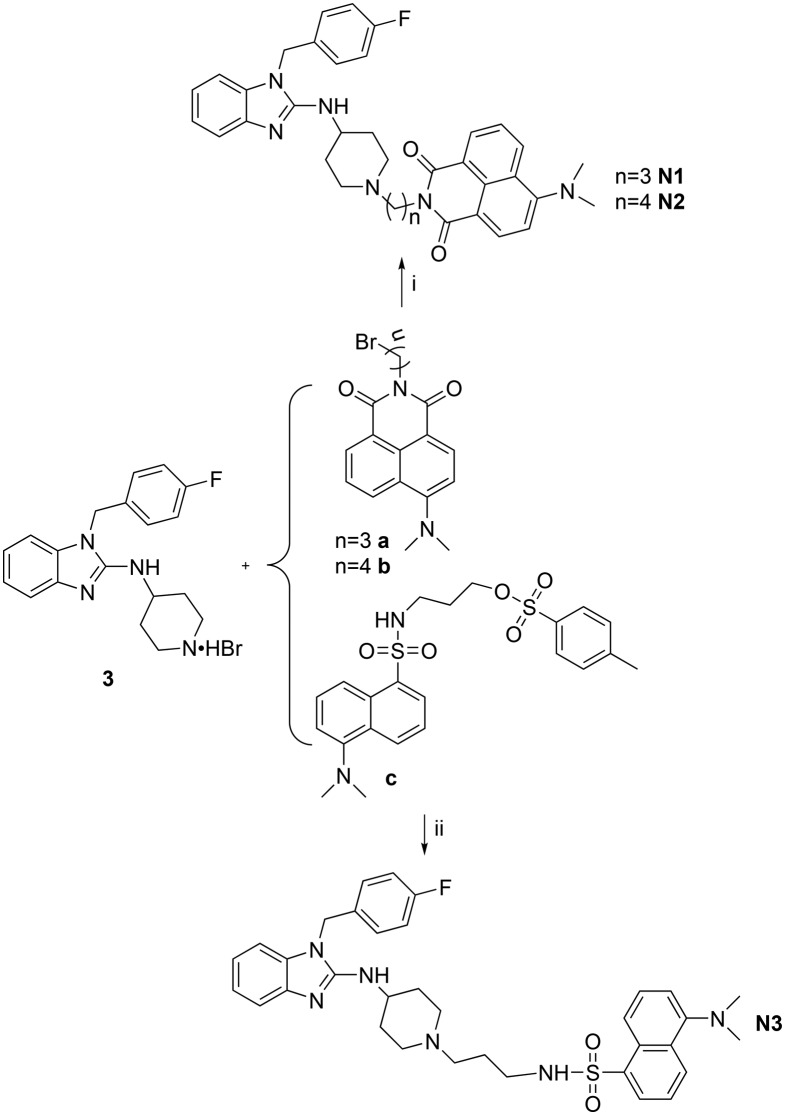
Subsequently, the fluorescent derivatives of astemizole were synthesized through a convenient reaction as shown in Scheme 2. Compound 3 and a (b, c) in the presence of K2CO3 in acetonitrile yielded the fluorescent compound N1 (N2, N3) through a substitution reaction. Further details about the synthesis of the fluorophores and recognition moiety can be found in the ESI.†
Scheme 2. Synthetic routes of the fluorescent probes. (i) K2CO3, acetonitrile, 80 °C; (ii) K2CO3, acetonitrile, 80 °C.
Spectroscopic properties of the probes
The spectroscopic properties of the probes were measured in a 10 μM solution of the corresponding probe (N1, N2, N3) in PBS (pH = 7.4). The results indicated that all probes possessed good fluorescent properties. Compared to probe N3, the maximum emissions of the probes N1 and N2 were red-shifted reaching 535 and 545 nm, respectively, but the fluorescence quantum yields were relatively low (Table 1).
Table 1. Photophysical properties of synthesized probes.
| Compd | λ max | λ ex | λ em | Φ (%) |
| N1 | 440 | 440 | 535 | 5 |
| N2 | 445 | 430 | 545 | 5 |
| N3 | 330 | 335 | 495 | 15 |
Binding affinity of probes
Soon afterwards, the inhibitory activities of these probes against the hERG channel were measured by radio-ligand binding assays using hERG transfected HEK293 cells.20–22 The results showed that probe N1 displayed the best inhibitory effects against the hERG channel, and the calculated IC50 and Ki values were 0.053 and 0.030 μM, respectively, which are slightly higher than astemizole (0.011 and 0.006 μM, respectively). Probes N2 and N3 also displayed potent inhibitory activities against the hERG channel, although lower than that of probe N1, with IC50 values of 0.183 and 0.186 μM, respectively (Table 2).
Table 2. Inhibitory activities of the synthesized probes against the hERG potassium channel.
aSee ESI.
bThe inhibition constant (Ki) was calculated from each IC50 value using the Cheng–Prusoff equation.
Cytotoxicity assay
The cytotoxicity of these probes was evaluated by CCK-8 assays using hERG transfected HEK293 cells. The results demonstrated that the IC50 of probes N1–N3 were 3.55 ± 0.28, 2.43 ± 0.12, and 7.03 ± 0.14 μM, respectively, in hERG–HEK293 cells (Table 3).
Table 3. Cytotoxicity results for the synthesized probes.
| Compd | IC50 (μM) |
| N1 | 3.55 ± 0.28 |
| N2 | 2.43 ± 0.12 |
| N3 | 7.03 ± 0.14 |
| Astemizole | 17.37 ± 1.07 |
Fluorescent image assay
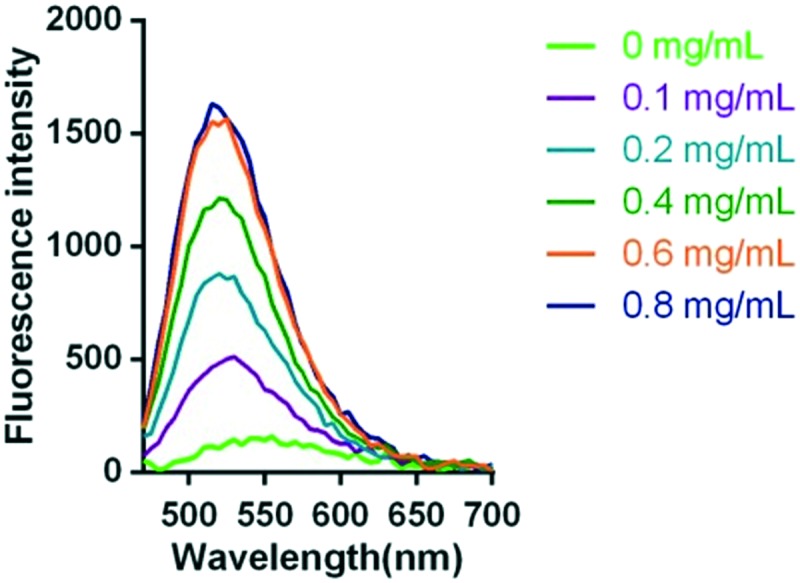
As mentioned above, the pharmacophore is environment-sensitive so the probe was proposed to have a turn-on mechanism for hERG channel. To test this hypothesis, a series of concentrations of hERG transfected HEK293 cell membranes were incubated with the probe N1 (5 μM). As good as anticipated, with an increase in the amounts of cell membranes, fluorescence intensity was gradually enhanced (Fig. 1). When incubated with 0.8 mg mL–1 cell membrane, the fluorescence intensity was 12-fold higher than that of the blank group.
Fig. 1. Fluorescent emission spectra of 5 μM probe N1 incubated with different concentrations of hERG transfected HEK293 membrane (0.8, 0.6, 0.4, 0.2, 0.1, and 0 mg mL–1) for 20–30 min in the assay buffer (50 mM Tris-HCl, 1 mM MgCl2, 10 mM KCl) at room temperature (λex = 440 nm).
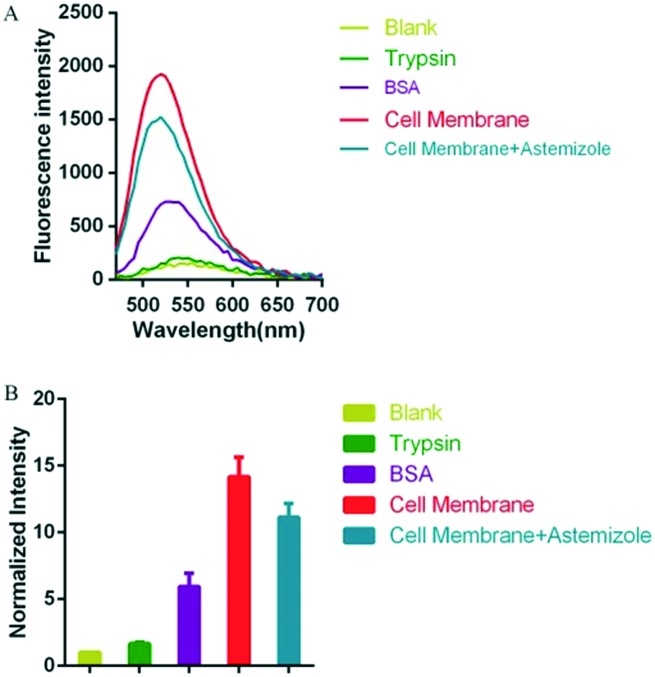
Subsequently, the selectivity of fluorescence intensity for hERG potassium channel was also examined. In the assay, taking into account the occurrence of nonspecific binding with small molecules, trypsin and bovine serum albumin (BSA) were selected as the control groups. Probe N1 (5 μM) was incubated with trypsin, BSA or hERG transfected HEK293 cell membrane at the same concentration (1 mg mL–1). As shown in Fig. 2, there is a suitable increase in the intensity for trypsin and BSA, which manifested that probe N1 may form some nonspecific binding with trypsin and BSA. Additionally, when probe N1 (5 μM) was incubated with the cell membrane (1 mg mL–1) and astemizole (a potent hERG channel inhibitor, 25 μM), the fluorescence intensity decreased compared with that of the group that was only incubated with the cell membrane. However, the degree of decrease in the fluorescence intensity was not complete, which may be caused by unavoidable nonspecific binding between probe N1 and other components in the cell membrane, especially the hydrophobic components.
Fig. 2. (A) Fluorescent emission spectra of 5 μM probe N1, which is respectively incubated with 1 mg mL–1 trypsin, 1 mg mL–1 BSA, 1 mg mL–1 hERG transfected HEK293 membrane, and 1 mg mL–1 cell membrane combined with hERG channel inhibitor astemizole (10 μM) for 20–30 min in the assay buffer (50 mM Tris-HCl, 1 mM MgCl2, 10 mM KCl) at room temperature (λex = 440 nm). (B) The corresponding fluorescence intensity changes (normalized based on the last point that is seen as 1) at 535 nm (λex = 440 nm).
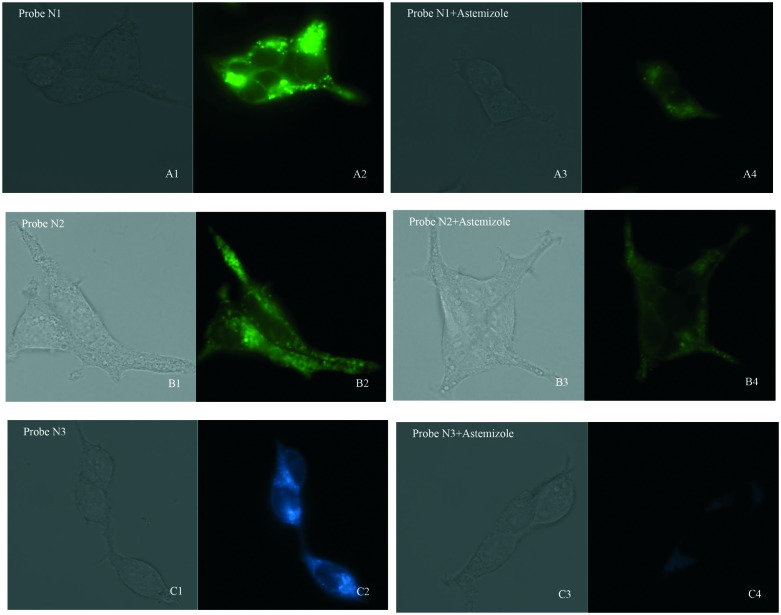
On account of their good fluorescent properties, acceptable cell toxicity, and potent inhibitory activity, probes N1–N3 can be used for hERG channel imaging in living cells in order to expand the application of our probes. Thus, microscopic imaging of probes N1–N3 for hERG channels in living cells was conducted on hERG transfected HEK293 cells. The microscopic imaging results indicated that these probes exhibit rapid responses and strong fluorescence toward hERG–HEK293 cells (Fig. 3). Astemizole, a potent inhibitor of the hERG channels, was chosen to be incubated with the cells and each of the probes. When the cells were coincubated with astemizole and the probes, the microscopic imaging results showed that the inhibition of hERG by astemizole resulted in a decrease in the fluorescence intensity, which confirmed that the hERG channel can be selectively labeled by the probe. Particularly, because of their turn-on mechanism, there was no need for a complex washing procedure, which ensured a convenient imaging process.23,24 Additionally, we investigated cell autofluorescence and the effect of astemizole on cell autofluorescence (see ESI†). The microscopic imaging results showed that the autofluorescence of the cells is negligible in both the presence and absence of astemizole (Fig. S9†), therefore, it would not interfere with the imaging of cells using probes N1–N3. In general, the results showed that these probes all possess superior selectivity toward hERG channels and could be used in the detection of hERG potassium channel.
Fig. 3. Fluorescence microscopy imaging of hERG transfected HEK293 cells incubated with 0.3 μM probe N1 (A1, bright field; A2, GFP channel), 0.3 μM probe N2 (B1, bright field; B2, GFP channel), and 0.5 μM probe N3 (C1, bright field; C2, DAPI channel), respectively. The imaging of inhibition of the hERG channels was accomplished by incubating astemizole (3 μM; 3 μM; 5 μM) with probe N1 (0.3 μM; A3, bright field; A4, GFP channel), N2 (0.3 μM; B3, bright field; B4, GFP channel), and N3 (0.5 μM; C3, bright field; C4, DAPI channel). All cells were incubated with each probe at 37 °C for 10 min and washed immediately. The background was adjusted by ImageJ software. Imaging was performed using a Zeiss Axio Observer A1 microscope with a 63× objective lens.
Conclusions
In conclusion, we developed three high-affinity environment-sensitive probes with commendable fluorescence properties, which can be utilized in the localization and visualization of hERG channels. These probes have been successfully utilized to label the hERG channels in hERG transfected HEK293 (hERG–HEK293) cells at the nanomolar level. Because of the environment-sensitive switch in the structure, there is no need for a washing procedure. Hence, this method is convenient compared with other imaging techniques, such as fluorescent protein-based approaches and immuno-fluorescence. Moreover, the synthesis of these probes is convenient and affordable. When compared with our previously published astemizole-based fluorescent probes,23,24 these probes have better turn-on effect and higher fluorescence intensity. They also have good fluorescence properties, acceptable cell toxicity, and potent inhibitory activity, which make the probes favourable for cell imaging. Therefore, these probes are anticipated to be applied in the studies of hERG channels. However, there are still a lot of work to be done, such as setting a screening system for hERG channels.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
The present project was supported by grants from the Shandong Natural Science Foundation (No. ZR2017MH101) and the Key Research and Development Project of Shandong Province (No. 2017CXGC1401).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c8md00562a
References
- Sanguinetti M. C., Tristani-Firouzi M. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- Roden D. M., Viswanathan P. C. J. Clin. Invest. 2005;115:2025–2032. doi: 10.1172/JCI25539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan M. D., Roden M. N. Engl. J. Med. 2004;350:1013–1022. doi: 10.1056/NEJMra032426. [DOI] [PubMed] [Google Scholar]
- Du L., Li M., You Q., Xia L. Biochem. Biophys. Res. Commun. 2007;355:889–894. doi: 10.1016/j.bbrc.2007.02.068. [DOI] [PubMed] [Google Scholar]
- Wang W., MacKinnon R., Cell, 2017, 169 , 422 –430 , , e410 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini A., Crociani O. Br. J. Cancer. 2000;83:1722–1729. doi: 10.1054/bjoc.2000.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. A., Tsui H. W., Newell E. W., Jiang X., Zhu X. P., Tsui F. W., Schlichter L. C. J. Biol. Chem. 2002;277:18528–18534. doi: 10.1074/jbc.M200592200. [DOI] [PubMed] [Google Scholar]
- Chen S. Z., Jiang M., Zhen Y. S. Cancer Chemother. Pharmacol. 2005;56:212–220. doi: 10.1007/s00280-004-0960-5. [DOI] [PubMed] [Google Scholar]
- Jehle J., Schweizer P. A., Katus H. A., Thomas D. Cell Death Dis. 2011;2:e193. doi: 10.1038/cddis.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin V. S., Chang C. J. Curr. Opin. Chem. Biol. 2012;16:595–601. doi: 10.1016/j.cbpa.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Tian X., Shin I., Yoon J. Chem. Soc. Rev. 2011;40:4783–4804. doi: 10.1039/c1cs15037e. [DOI] [PubMed] [Google Scholar]
- Lin V. S., Lippert A. R., Chang C. J. Proc. Natl. Acad. Sci. U. S. A. 2013;110:7131–7135. doi: 10.1073/pnas.1302193110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov A. Drug Discovery Today. 2005;10:149–155. doi: 10.1016/S1359-6446(04)03278-7. [DOI] [PubMed] [Google Scholar]
- Yamakawa Y., Furutani K., Inanobe A., Ohno Y., Kurachi Y. Biochem. Biophys. Res. Commun. 2012;418:161–166. doi: 10.1016/j.bbrc.2011.12.153. [DOI] [PubMed] [Google Scholar]
- Cohen B. E., McAnaney T. B., Park E. S., Jan Y. N., Boxer S. G., Jan L. Y. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- Barbara I., Loving G. Bioconjugate Chem. 2009;20:2133–2141. doi: 10.1021/bc900319z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexei Toutchkine W. H., Ullmann M. J. Phys. Chem. A. 2007;111:10849–10860. doi: 10.1021/jp073197r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayab P. S., Pulaganti M., Chitta S. K., Abid M., Uddin R. J. Fluoresc. 2015;25:1905–1920. doi: 10.1007/s10895-015-1683-1. [DOI] [PubMed] [Google Scholar]
- Ozdemir E., Tatar Ulu S. Luminescence. 2017;32:1145–1149. doi: 10.1002/bio.3301. [DOI] [PubMed] [Google Scholar]
- Diaz G. J., Daniell K., Leitza S. T., Martin R. L., Su Z., McDermott J. S., Cox B. F., Gintant G. A. J. Pharmacol. Toxicol. Methods. 2004;50:187–199. doi: 10.1016/j.vascn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Huang X. P., Mangano T., Hufeisen S., Setola V., Roth B. L. Assay Drug Dev. Technol. 2010;8:727–742. doi: 10.1089/adt.2010.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus S. A., Beacham D., Shahane S. A., Southall N., Xia M., Huang R., Hooten E., Zhao Y., Shou L., Austin C. P., Zheng W. Anal. Biochem. 2009;394:30–38. doi: 10.1016/j.ab.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Liu Z., Ma Z., Li M., Du L. ACS Med. Chem. Lett. 2016;7:245–249. doi: 10.1021/acsmedchemlett.5b00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Jiang T., Wang B., Ke B., Zhou Y., Du L., Li M. Anal. Chem. 2016;88:1511–1515. doi: 10.1021/acs.analchem.5b04220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.